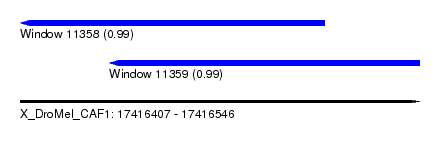
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,416,407 – 17,416,546 |
| Length | 139 |
| Max. P | 0.989992 |

| Location | 17,416,407 – 17,416,513 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -23.67 |
| Energy contribution | -23.34 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17416407 106 - 22224390 GUGUUUAACUAUUGGGCUACCAAUCGCAAUUUGUCAGGGCCCAGAAUCCAAGGGCCAA--GGC-GGGGGAAACCCAAUUCGAUGACUUCAAUUAUUGGAUGUAUGUAAC (((.......(((((....))))))))....((((..(((((.........)))))..--)))-)(((....))).(((((((((......)))))))))......... ( -32.01) >DroEre_CAF1 10029 103 - 1 GUAUUCCACUAUUGGGUUACCAAUCGCAAUUUGUCAAGGCCCAGAAUAAAAGGGCUAAAA--CAGGGGGAAACCCAAUGCGAUAACUUCAAUUAUUAA----AUGUAAC (((..(((....)))..)))..((((((..((((...(((((.........)))))...)--)))(((....)))..))))))...............----....... ( -29.40) >DroYak_CAF1 9966 96 - 1 GUGUUCAACUAUUGGGUUACCAAUCACAAUUUGUCAGGGCCCAGAAUCAAAGGGCUCAAA--C-GGGGGAAACCCAAUUCGAUGACAUCAAUUAUU----------GAC ....((((..(((((....))))).......(((((((((((.........))))))...--.-.(((....))).......))))).......))----------)). ( -29.10) >consensus GUGUUCAACUAUUGGGUUACCAAUCGCAAUUUGUCAGGGCCCAGAAUCAAAGGGCUAAAA__C_GGGGGAAACCCAAUUCGAUGACUUCAAUUAUU______AUGUAAC (((.......(((((....))))))))...(((((..(((((.........))))).........(((....))).....)))))........................ (-23.67 = -23.34 + -0.33)


| Location | 17,416,438 – 17,416,546 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -29.03 |
| Energy contribution | -28.49 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17416438 108 - 22224390 -------UCUGACAUUGGCCUAAAUUCAAAUAUGAUUUUUGUGUUUAACUAUUGGGCUACCAAUCGCAAUUUGUCAGGGCCCAGAAUCCAAGGGCCAA--GGC-GGGGGAAACCCAAU -------.((((((...((.(((((.((((.......)))).)))))...(((((....))))).))....))))))(((((.........)))))..--...-.(((....)))... ( -35.70) >DroEre_CAF1 10056 116 - 1 UCCCUGAUCUGACAUUGGCCUAAAUUCAAAUACGAUUUUUGUAUUCCACUAUUGGGUUACCAAUCGCAAUUUGUCAAGGCCCAGAAUAAAAGGGCUAAAA--CAGGGGGAAACCCAAU .((((..((((.....(((((.....(((((.(((((...(((..(((....)))..))).)))))..)))))...))))))))).....))))......--...(((....)))... ( -33.60) >DroYak_CAF1 9987 105 - 1 -------UCUGACAUUGGCCUAAAUUCAAAUAC---UUCUGUGUUCAACUAUUGGGUUACCAAUCACAAUUUGUCAGGGCCCAGAAUCAAAGGGCUCAAA--C-GGGGGAAACCCAAU -------..(((((.((((((((.....(((((---....)))))......))))))))............)))))((((((.........))))))...--.-.(((....)))... ( -30.12) >consensus _______UCUGACAUUGGCCUAAAUUCAAAUACGAUUUUUGUGUUCAACUAUUGGGUUACCAAUCGCAAUUUGUCAGGGCCCAGAAUCAAAGGGCUAAAA__C_GGGGGAAACCCAAU ........((((((.((((((((.....(((((.......)))))......))))))))............))))))(((((.........))))).........(((....)))... (-29.03 = -28.49 + -0.55)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:35 2006