
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,415,235 – 17,415,364 |
| Length | 129 |
| Max. P | 0.979726 |

| Location | 17,415,235 – 17,415,338 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -12.87 |
| Energy contribution | -12.76 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17415235 103 + 22224390 UCUCUU-GGCC-CGGUGUUU------UGCUAUUAU--UUUCUUAACAGUUAGCGGGCAGCAUACUG---UUUUUUUGUAUAAUU--U--UUUAAUGCCUUUCCUAACUGGAUUGGCAAUU ......-.(((-((.((...------.(((.(((.--.....))).))))).)))))...((((..---.......))))....--.--.....((((..(((.....)))..))))... ( -24.40) >DroSec_CAF1 8908 104 + 1 UCUCUU-GGCC-CGGUGUUU------UGCUAUUAU--UUUCUUUACAGUUAGCGGGCAGCAUACUG---UUU-UUUGUAUACUU--UUUUUUAAUGCCUUUCCUAACUGGAUUGGCAAUU ......-.(((-((.((..(------((.((....--......))))).)).)))))((.((((..---...-...)))).)).--........((((..(((.....)))..))))... ( -23.70) >DroSim_CAF1 8984 104 + 1 UCUCUU-GGCC-CGAUGUUU------UGCUAUUAU--UUUCUUUACAGUUAGCGGGCAGCAUACUG---UUU-UUUGUAUAAUU--UUUUUUAAUGCCUUUCCUAACUGGAUUGGCAAUU ......-.(((-((.(((..------.........--.......))).....)))))...((((..---...-...))))....--........((((..(((.....)))..))))... ( -22.47) >DroEre_CAF1 8809 104 + 1 UCUCUU-CGCC-CGGUGUUC------UGCUAUUAU--UUUCUAUACAGUUAUCGAGCAGCUUACUGUGGUUU--UUUUAUAAUU----UUUUAAUGCCUUUCUUAACUGGAUUGGCAAUU ......-.(((-(((((..(------((((.....--...((....))......)))))..))))).)))..--..........----......((((..(((.....)))..))))... ( -23.84) >DroYak_CAF1 8740 106 + 1 UCUCGG-CGCCCCGGUGUUU------UGCUAUUAU--UUUCUUUACAGUUAGCGAGCAGCUUACUG---UUU--UUUUAUACUUUUUUUUUUAAUGCCUUUCUUAACUGGAUUGGCAAUU ....((-(((....))))).------.........--.......(((((.(((.....))).))))---)..--....................((((..(((.....)))..))))... ( -21.80) >DroAna_CAF1 8800 99 + 1 UCUAUUUGGCC-CGCUUUACCAUUAUUAUUAUUAUUUUUUCUUUACAGUUAGUGAGCAGAGUAGUG---UUG-C--------UU--U------UUGCCUUUCUUAACUGGAUUGGCAAUU .((((((.((.-((((..................................)))).)).))))))..---...-.--------..--.------(((((..(((.....)))..))))).. ( -17.98) >consensus UCUCUU_GGCC_CGGUGUUU______UGCUAUUAU__UUUCUUUACAGUUAGCGAGCAGCAUACUG___UUU_UUUGUAUAAUU__U_UUUUAAUGCCUUUCCUAACUGGAUUGGCAAUU ............((((((........(((((..................)))))......))))))............................((((..(((.....)))..))))... (-12.87 = -12.76 + -0.11)
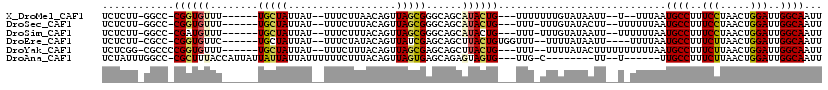

| Location | 17,415,265 – 17,415,364 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -16.67 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17415265 99 + 22224390 CUUAACAGUUAGCGGGCAGCAUACUG---UUUUUUUGUAUAAUU--U--UUUAAUGCCUUUCCUAACUGGAUUGGCAAUUG--------CG------CCUGGCUGUAUAUUAAAUCAUUU ....((((((((.(.((((.((((..---.......))))....--.--.....((((..(((.....)))..)))).)))--------).------))))))))).............. ( -28.10) >DroSec_CAF1 8938 98 + 1 CUUUACAGUUAGCGGGCAGCAUACUG---UUU-UUUGUAUACUU--UUUUUUAAUGCCUUUCCUAACUGGAUUGGCAAUUG----------------CCUGGCUGUAUAUUAAAUCAUUU ...((((((...(((((((.((((..---...-...))))....--........((((..(((.....)))..)))).)))----------------))))))))))............. ( -28.30) >DroSim_CAF1 9014 98 + 1 CUUUACAGUUAGCGGGCAGCAUACUG---UUU-UUUGUAUAAUU--UUUUUUAAUGCCUUUCCUAACUGGAUUGGCAAUUG----------------CCUGGCUGUAUAUUAAAUCAUUU ...((((((...(((((((.((((..---...-...))))....--........((((..(((.....)))..)))).)))----------------))))))))))............. ( -28.30) >DroEre_CAF1 8839 98 + 1 CUAUACAGUUAUCGAGCAGCUUACUGUGGUUU--UUUUAUAAUU----UUUUAAUGCCUUUCUUAACUGGAUUGGCAAUUG----------------CCUGGCUGUAUAUUAAAUCAUUU .((((((((((....((((....))))(((..--.....(((..----..))).((((..(((.....)))..))))...)----------------))))))))))))........... ( -22.10) >DroYak_CAF1 8771 99 + 1 CUUUACAGUUAGCGAGCAGCUUACUG---UUU--UUUUAUACUUUUUUUUUUAAUGCCUUUCUUAACUGGAUUGGCAAUUG----------------CCUGGCUGUAUAUUAAAUCAUUU ...((((((((((((((((....)))---)))--....................((((..(((.....)))..))))...)----------------.)))))))))............. ( -23.80) >DroAna_CAF1 8839 100 + 1 CUUUACAGUUAGUGAGCAGAGUAGUG---UUG-C--------UU--U------UUGCCUUUCUUAACUGGAUUGGCAAUUGGUGUGCUGCGCUACUAACUGGUCGCAUAUUAAAUCAUUU .....((((((((.(((...((((..---(.(-(--------(.--.------(((((..(((.....)))..)))))..))))..)))))))))))))))................... ( -32.60) >consensus CUUUACAGUUAGCGAGCAGCAUACUG___UUU_UUUGUAUAAUU__U_UUUUAAUGCCUUUCCUAACUGGAUUGGCAAUUG________________CCUGGCUGUAUAUUAAAUCAUUU ...(((((((((....(((....)))............................((((..(((.....)))..)))).....................)))))))))............. (-16.67 = -17.12 + 0.44)
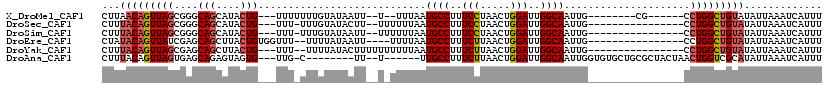


| Location | 17,415,265 – 17,415,364 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -19.45 |
| Consensus MFE | -9.39 |
| Energy contribution | -9.62 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17415265 99 - 22224390 AAAUGAUUUAAUAUACAGCCAGG------CG--------CAAUUGCCAAUCCAGUUAGGAAAGGCAUUAAA--A--AAUUAUACAAAAAAA---CAGUAUGCUGCCCGCUAACUGUUAAG .......((((((...(((..((------((--------(...((((..(((.....)))..)))).....--.--....((((.......---..)))))).))).)))...)))))). ( -20.50) >DroSec_CAF1 8938 98 - 1 AAAUGAUUUAAUAUACAGCCAGG----------------CAAUUGCCAAUCCAGUUAGGAAAGGCAUUAAAAAA--AAGUAUACAAA-AAA---CAGUAUGCUGCCCGCUAACUGUAAAG .............(((((.(.((----------------(...((((..(((.....)))..))))........--.(((((((...-...---..)))))))))).)....)))))... ( -23.70) >DroSim_CAF1 9014 98 - 1 AAAUGAUUUAAUAUACAGCCAGG----------------CAAUUGCCAAUCCAGUUAGGAAAGGCAUUAAAAAA--AAUUAUACAAA-AAA---CAGUAUGCUGCCCGCUAACUGUAAAG ..(((((((........(((.((----------------(....)))..(((.....)))..)))........)--)))))).....-..(---((((..((.....))..))))).... ( -20.19) >DroEre_CAF1 8839 98 - 1 AAAUGAUUUAAUAUACAGCCAGG----------------CAAUUGCCAAUCCAGUUAAGAAAGGCAUUAAAA----AAUUAUAAAA--AAACCACAGUAAGCUGCUCGAUAACUGUAUAG ...........(((((((((.((----------------(....)))..((.......))..))).......----..........--......(((....))).........)))))). ( -14.90) >DroYak_CAF1 8771 99 - 1 AAAUGAUUUAAUAUACAGCCAGG----------------CAAUUGCCAAUCCAGUUAAGAAAGGCAUUAAAAAAAAAAGUAUAAAA--AAA---CAGUAAGCUGCUCGCUAACUGUAAAG ...........(((((.(((.((----------------(....)))..((.......))..))).((......))..)))))...--..(---((((.(((.....))).))))).... ( -17.90) >DroAna_CAF1 8839 100 - 1 AAAUGAUUUAAUAUGCGACCAGUUAGUAGCGCAGCACACCAAUUGCCAAUCCAGUUAAGAAAGGCAA------A--AA--------G-CAA---CACUACUCUGCUCACUAACUGUAAAG ...................((((((((.(.((((........(((((..((.......))..)))))------.--.(--------(-...---..))...)))).)))))))))..... ( -19.50) >consensus AAAUGAUUUAAUAUACAGCCAGG________________CAAUUGCCAAUCCAGUUAAGAAAGGCAUUAAAA_A__AAUUAUACAAA_AAA___CAGUAUGCUGCCCGCUAACUGUAAAG .............(((((.........................((((..((.......))..))))............................(((....)))........)))))... ( -9.39 = -9.62 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:34 2006