
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,413,870 – 17,413,967 |
| Length | 97 |
| Max. P | 0.936342 |

| Location | 17,413,870 – 17,413,967 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 89.34 |
| Mean single sequence MFE | -27.95 |
| Consensus MFE | -22.00 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17413870 97 + 22224390 GUGUGUGUGAAUGUGUUUGGCGUGCGGCUAACACUGUU---GUUGUCGAAUUGUUGUUGUUGUUGGUAAAAACUUGCGCGCCAUUACUAACAGUAAAUUA .....(((....(((..(((((((((((.((((....)---))))))..............(((......)))..)))))))).)))..)))........ ( -23.40) >DroSec_CAF1 7504 97 + 1 GUGUGUGUGAGUGUGUUUGGCGUGCGGCUAACACUGUU---GUUGUCCAAUUGUUGUUGUUGUUGGUAACAACUUGCGCGCCAUUACUAACAGUAAAUUG .....(((.((((....((((((((((.(((((....)---)))).)).......(((((((....)))))))..)))))))).)))).)))........ ( -31.00) >DroSim_CAF1 7578 97 + 1 GUGUGUGUGAGUGUGUUUGACGUGCGGCUAACACUGUU---GUUGUCCAAUUGUUGUUGUUGUUGGUAACAACUUGCGCGCCAUUACUAACAGUAAAUUG (.(((((..(((((..(..(((.(((((.((((..(((---(.....))))))))))))))))..)..)).)))..)))))).((((.....)))).... ( -26.00) >DroYak_CAF1 7457 94 + 1 GUGUGU------GUGUUUGGCGUGCGGCUAACUCCGUUGUUGUUGCCCAAUUGUAGUUGUUGUUGGUAACAACGUGCGCGCCAUUAUUAACAGUAACUUA ...(((------.....((((((((((......))...(((((((((((((.......))))..)))))))))..))))))))......)))........ ( -31.40) >consensus GUGUGUGUGAGUGUGUUUGGCGUGCGGCUAACACUGUU___GUUGUCCAAUUGUUGUUGUUGUUGGUAACAACUUGCGCGCCAUUACUAACAGUAAAUUA ...(((......(((..(((((((((((.(((.........))))))........(((((((....)))))))..)))))))).)))..)))........ (-22.00 = -22.12 + 0.12)

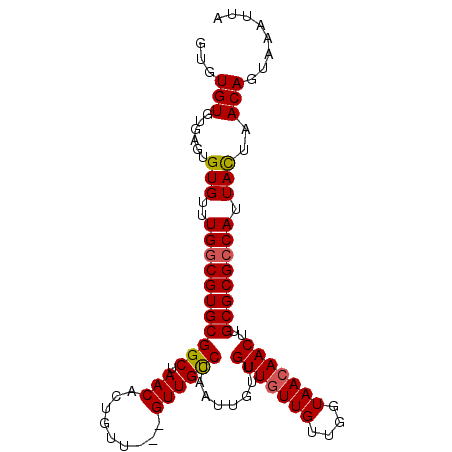
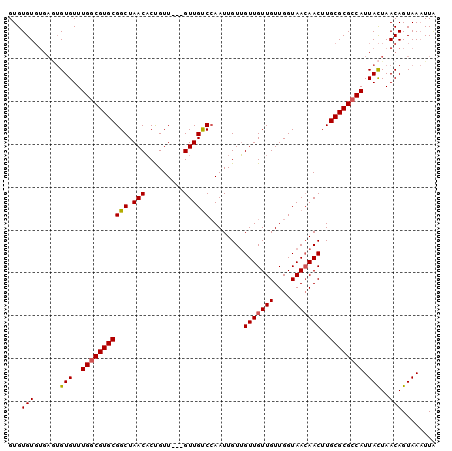
| Location | 17,413,870 – 17,413,967 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.34 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -15.66 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17413870 97 - 22224390 UAAUUUACUGUUAGUAAUGGCGCGCAAGUUUUUACCAACAACAACAACAAUUCGACAAC---AACAGUGUUAGCCGCACGCCAAACACAUUCACACACAC ....((((.....))))(((((.((..(((......)))..............((((..---.....))))....)).)))))................. ( -14.00) >DroSec_CAF1 7504 97 - 1 CAAUUUACUGUUAGUAAUGGCGCGCAAGUUGUUACCAACAACAACAACAAUUGGACAAC---AACAGUGUUAGCCGCACGCCAAACACACUCACACACAC ........(((.(((..(((((.((..((((((......)))))).......((..(((---(....))))..)))).))))).....))).)))..... ( -23.60) >DroSim_CAF1 7578 97 - 1 CAAUUUACUGUUAGUAAUGGCGCGCAAGUUGUUACCAACAACAACAACAAUUGGACAAC---AACAGUGUUAGCCGCACGUCAAACACACUCACACACAC ........(((.(((..(((((.((..((((((......)))))).......((..(((---(....))))..)))).))))).....))).)))..... ( -20.90) >DroYak_CAF1 7457 94 - 1 UAAGUUACUGUUAAUAAUGGCGCGCACGUUGUUACCAACAACAACUACAAUUGGGCAACAACAACGGAGUUAGCCGCACGCCAAACAC------ACACAC ...((...((((.....(((((.((..((((((.((((............))))..))))))...((......)))).))))))))).------..)).. ( -23.10) >consensus CAAUUUACUGUUAGUAAUGGCGCGCAAGUUGUUACCAACAACAACAACAAUUGGACAAC___AACAGUGUUAGCCGCACGCCAAACACACUCACACACAC .................(((((.((..((((((......)))))).......((..(((.........)))..)))).)))))................. (-15.66 = -15.98 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:31 2006