
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,380,335 – 17,380,450 |
| Length | 115 |
| Max. P | 0.981317 |

| Location | 17,380,335 – 17,380,430 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -23.90 |
| Energy contribution | -23.28 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
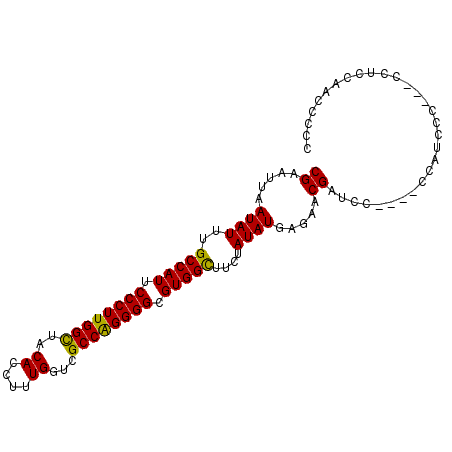
>X_DroMel_CAF1 17380335 95 + 22224390 CGAAUUAAUAUUUGCCAUUCCCUUGGUUACACCUUUGGUCGCCAGGGGCGUGGCUUCUAUAUGAGAACGAUCC----UCAUCCCCAGCCUGCAACGCGC .............(((((.((((((((.((.......)).)))))))).)))))......(((((.......)----)))).....((.......)).. ( -26.90) >DroSec_CAF1 97222 96 + 1 CGAAUUAAUAUUUGCCAUUCCCUUGGCUACACAUUUGGUCGCCAGGGGCGUGGCUUCUAUAUGAGAACGAUCCCAUCCCAUCCC---CCUGCAACGCGC ((.....((((..(((((.((((((((.((.(....))).)))))))).)))))....)))).....))...............---............ ( -26.60) >DroEre_CAF1 97963 82 + 1 CGAAUUAAUAUUUGCCAUUCCCUUGGCUACACCUUUGCUCGCCAGGGGCGUGGUUUUCAUAUGAGGACGAUCC-----------------CCAUGCCCC .............(((((.((((((((.............)))))))).))))).....((((.((.....))-----------------.)))).... ( -24.72) >DroYak_CAF1 89373 93 + 1 CGAAUUAAUAUUUGCCAUUCCCUUGGCUACACCUUUGUUGGCCGGGGGCGUGGUCUCUAUAUGAGGGCGAUCCU---CGAUUCC---CCUCCAAUCCCC .......((((..(((((.((((((((((((....)).)))))))))).)))))....))))(((((.((((..---.)))).)---))))........ ( -33.90) >consensus CGAAUUAAUAUUUGCCAUUCCCUUGGCUACACCUUUGGUCGCCAGGGGCGUGGCUUCUAUAUGAGAACGAUCC____CCAUCCC___CCUCCAACCCCC ((.....((((..(((((.((((((((..((....))...)))))))).)))))....)))).....)).............................. (-23.90 = -23.28 + -0.62)

| Location | 17,380,356 – 17,380,450 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 74.69 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -13.61 |
| Energy contribution | -13.05 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17380356 94 + 22224390 CUUGGUUACACCUUUGGUCGCCAGGGGCGUGGCUUCUAUAUGAGAACGAUCC----UCAUCCCCAGCCUGCAACGCGCUUCAUCCCCACUAUCCCCCC ..((((.((.......)).))))(((((((((((.....(((((.......)----))))....))))......)))))))................. ( -21.60) >DroSec_CAF1 97243 95 + 1 CUUGGCUACACAUUUGGUCGCCAGGGGCGUGGCUUCUAUAUGAGAACGAUCCCAUCCCAUCCC---CCUGCAACGCGCUUCAUCCCCACUAUCAUCCC ...(((((......)))))((((((((.((((.((((.....)))).(((...))))))).))---))))....))...................... ( -20.20) >DroEre_CAF1 97984 79 + 1 CUUGGCUACACCUUUGCUCGCCAGGGGCGUGGUUUUCAUAUGAGGACGAUCC-----------------CCAUGCCCCACCAUCACCACUAUCCCC-- ..((((.............))))(((((((((...((....))((.....))-----------------)))))))))..................-- ( -23.52) >DroYak_CAF1 89394 92 + 1 CUUGGCUACACCUUUGUUGGCCGGGGGCGUGGUCUCUAUAUGAGGGCGAUCCU---CGAUUCC---CCUCCAAUCCCCACCAUCCCCACCAUCUCCCC ...(((((((....)).)))))(((((.(((((........(((((.((((..---.)))).)---))))........))))).))).))........ ( -31.29) >consensus CUUGGCUACACCUUUGGUCGCCAGGGGCGUGGCUUCUAUAUGAGAACGAUCC____CCAUCCC___CCUCCAACCCCCACCAUCCCCACUAUCCCCCC ...((((((.((((((.....)))))).))))))................................................................ (-13.61 = -13.05 + -0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:21 2006