
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,339,571 – 17,339,664 |
| Length | 93 |
| Max. P | 0.965611 |

| Location | 17,339,571 – 17,339,664 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.68 |
| Mean single sequence MFE | -19.54 |
| Consensus MFE | -12.60 |
| Energy contribution | -14.10 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
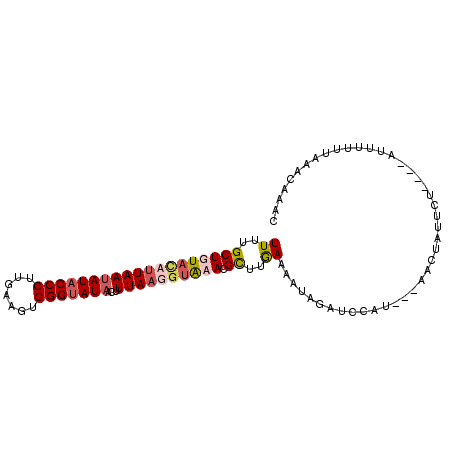
>X_DroMel_CAF1 17339571 93 + 22224390 UUUUGCUGUAAAUUAAUAUACCCUUGAAGUGGGUAUAACUUUAAGGUAAACGCUUGAAAAUAGGUUCAC---AACUAUUCU----UUUUUUUAAACAAAU .((((((.((((....(((((((.......)))))))..)))).)))))).(.(((((((.(((.....---.......))----).))))))).).... ( -16.00) >DroSec_CAF1 56800 94 + 1 UUUUGCUGUUUAUUAAUAUACCCUUGAAGUGGGUAUAACUUUAAGGUAAACGCUUGAAAAUAGGUUCA----AACCAUU--AUUAUUUUAUUAAACAAAU ......((((((.((((((((((.......))))))).......(((....(((((....)))))...----.)))...--......))).))))))... ( -17.80) >DroEre_CAF1 57372 96 + 1 UUUUGCUGUACAUUAAAAUACCCUUGAAAUGGGUAUAAUUUUAUUGUGCACGUUUAAAAAUAGAUCCAUCAUAGCUAUUCU----ACUUUUAAAUCAAAC ......((((((.((((((((((.......))))))...)))).)))))).((((((((.((((..............)))----).))))))))..... ( -20.44) >DroYak_CAF1 48168 96 + 1 UUUUGCUGUACAUUAAUAUACCCUUGAAGUGGGCAUAACUUUAAUGUGCACGCUUGAAAAUAGAUCCAUCAUAGCUAUUCU----AUUUUUGAAACAAAC ....(((((((((((((((.(((.......))).)))...)))))))))).))(..((((((((..............)))----)))))..)....... ( -23.94) >consensus UUUUGCUGUACAUUAAUAUACCCUUGAAGUGGGUAUAACUUUAAGGUAAACGCUUGAAAAUAGAUCCAU___AACUAUUCU____AUUUUUUAAACAAAC ((..(((((((((((((((((((.......)))))))...)))))))))).))..))........................................... (-12.60 = -14.10 + 1.50)

| Location | 17,339,571 – 17,339,664 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.68 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -12.88 |
| Energy contribution | -12.50 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
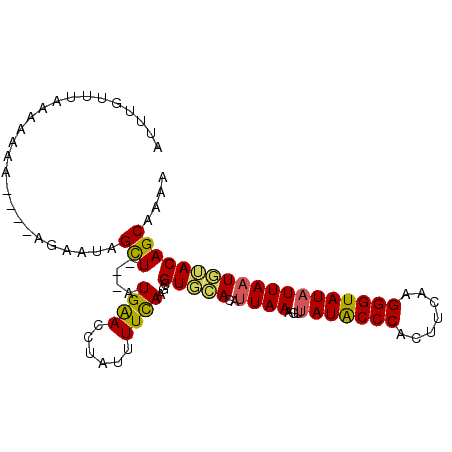
>X_DroMel_CAF1 17339571 93 - 22224390 AUUUGUUUAAAAAAA----AGAAUAGUU---GUGAACCUAUUUUCAAGCGUUUACCUUAAAGUUAUACCCACUUCAAGGGUAUAUUAAUUUACAGCAAAA .((((((((((....----.........---((((((((.......)).))))))........(((((((.......)))))))....)))).)))))). ( -16.80) >DroSec_CAF1 56800 94 - 1 AUUUGUUUAAUAAAAUAAU--AAUGGUU----UGAACCUAUUUUCAAGCGUUUACCUUAAAGUUAUACCCACUUCAAGGGUAUAUUAAUAAACAGCAAAA .((((((............--....(((----((((......)))))))(((((..((((...(((((((.......)))))))))))))))))))))). ( -19.30) >DroEre_CAF1 57372 96 - 1 GUUUGAUUUAAAAGU----AGAAUAGCUAUGAUGGAUCUAUUUUUAAACGUGCACAAUAAAAUUAUACCCAUUUCAAGGGUAUUUUAAUGUACAGCAAAA .((((.(((((((((----(((....((.....)).)))))))))))).(((((...((((...((((((.......)))))))))).)))))..)))). ( -20.50) >DroYak_CAF1 48168 96 - 1 GUUUGUUUCAAAAAU----AGAAUAGCUAUGAUGGAUCUAUUUUCAAGCGUGCACAUUAAAGUUAUGCCCACUUCAAGGGUAUAUUAAUGUACAGCAAAA (((((.....(((((----(((....((.....)).))))))))))))).((((((((((...(((((((.......))))))))))))))...)))... ( -21.70) >consensus AUUUGUUUAAAAAAA____AGAAUAGCU___AUGAACCUAUUUUCAAGCGUGCACAUUAAAGUUAUACCCACUUCAAGGGUAUAUUAAUGUACAGCAAAA .........................(((....((((......))))...(((((..((((...(((((((.......))))))))))))))))))).... (-12.88 = -12.50 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:09 2006