
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,308,273 – 17,308,365 |
| Length | 92 |
| Max. P | 0.796968 |

| Location | 17,308,273 – 17,308,365 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 93.71 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -17.40 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
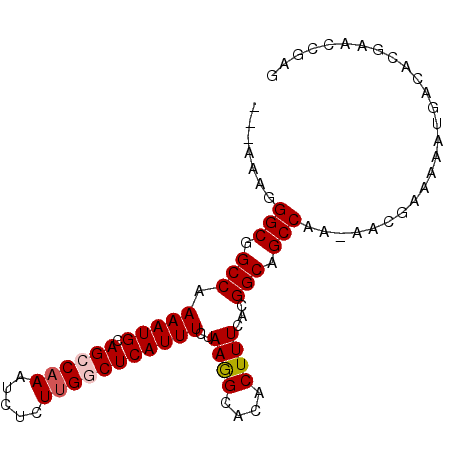
>X_DroMel_CAF1 17308273 92 + 22224390 AAAAAAAGGCGGCCAAAAUGCAGCCAAAUCUCUUGGCUCAUUUCUAAGGCACACUUUCACGGCAGCCAA-AACGAAAAAUGACACGAACCGAG .......(((.(((.(((((.((((((.....)))))))))))..(((.....)))....))).)))..-....................... ( -22.10) >DroSec_CAF1 25662 89 + 1 ---AAAGGGCGGCCAAAAUGCAGCCAAAUCUCUUGGCUCAUUUCUAAGGCACACUUUCACGGCAGCCAA-AACGAAAAAUGACACGAACCGAG ---....(((.(((.(((((.((((((.....)))))))))))..(((.....)))....))).)))..-....................... ( -21.90) >DroSim_CAF1 15361 89 + 1 ---AAAGGGCGGCCAAAAUGCAGCCAAAUCUCUUGGCUCAUUUCUAAGGCACACUUUCACGGCAGCCAA-AACGAAAAAUGACACGAACCGAG ---....(((.(((.(((((.((((((.....)))))))))))..(((.....)))....))).)))..-....................... ( -21.90) >DroEre_CAF1 26798 89 + 1 ---AAAGGGCGGCCCAAAUGCAGCCAAAUCUCUAGACUCAUUUCUAAAGCACACUUUCACGGCAGCCAA-AGCGAAAAAUGACACGAGCCGAG ---(((((((.((......)).)))....((.((((......)))).))....))))..((((.((...-.))(........)....)))).. ( -15.20) >DroYak_CAF1 16251 90 + 1 ---AAAGGGCGGCCAAAAUGCAGCCAAAUCUCUUGCCUCAUUUCUAAGGCACACUUUCACGGCAGCCAAAAGAGAAAAAUGACAUGAACCGAG ---....(((.((......)).)))...((((((((((........))))..........((...))..)))))).................. ( -17.90) >consensus ___AAAGGGCGGCCAAAAUGCAGCCAAAUCUCUUGGCUCAUUUCUAAGGCACACUUUCACGGCAGCCAA_AACGAAAAAUGACACGAACCGAG .......(((.(((.(((((.((((((.....)))))))))))..((((....))))...))).))).......................... (-17.40 = -17.84 + 0.44)


| Location | 17,308,273 – 17,308,365 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 93.71 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -23.18 |
| Energy contribution | -23.34 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17308273 92 - 22224390 CUCGGUUCGUGUCAUUUUUCGUU-UUGGCUGCCGUGAAAGUGUGCCUUAGAAAUGAGCCAAGAGAUUUGGCUGCAUUUUGGCCGCCUUUUUUU (.((((....((((.........-.)))).)))).)...(((.(((...(((((((((((((...))))))).))))))))))))........ ( -26.20) >DroSec_CAF1 25662 89 - 1 CUCGGUUCGUGUCAUUUUUCGUU-UUGGCUGCCGUGAAAGUGUGCCUUAGAAAUGAGCCAAGAGAUUUGGCUGCAUUUUGGCCGCCCUUU--- (.((((....((((.........-.)))).)))).)...(((.(((...(((((((((((((...))))))).)))))))))))).....--- ( -26.20) >DroSim_CAF1 15361 89 - 1 CUCGGUUCGUGUCAUUUUUCGUU-UUGGCUGCCGUGAAAGUGUGCCUUAGAAAUGAGCCAAGAGAUUUGGCUGCAUUUUGGCCGCCCUUU--- (.((((....((((.........-.)))).)))).)...(((.(((...(((((((((((((...))))))).)))))))))))).....--- ( -26.20) >DroEre_CAF1 26798 89 - 1 CUCGGCUCGUGUCAUUUUUCGCU-UUGGCUGCCGUGAAAGUGUGCUUUAGAAAUGAGUCUAGAGAUUUGGCUGCAUUUGGGCCGCCCUUU--- ..(((((((.((((.....((((-((.((....)).))))))..(((((((......)))))))...))))......)))))))......--- ( -25.60) >DroYak_CAF1 16251 90 - 1 CUCGGUUCAUGUCAUUUUUCUCUUUUGGCUGCCGUGAAAGUGUGCCUUAGAAAUGAGGCAAGAGAUUUGGCUGCAUUUUGGCCGCCCUUU--- ((((((....((((...........)))).))).........(((((((....))))))).)))....(((.((......)).)))....--- ( -26.40) >consensus CUCGGUUCGUGUCAUUUUUCGUU_UUGGCUGCCGUGAAAGUGUGCCUUAGAAAUGAGCCAAGAGAUUUGGCUGCAUUUUGGCCGCCCUUU___ (.((((....((((...........)))).)))).)...(((.(((...(((((((((((((...))))))).))))))))))))........ (-23.18 = -23.34 + 0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:53 2006