
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,294,303 – 17,294,433 |
| Length | 130 |
| Max. P | 0.911290 |

| Location | 17,294,303 – 17,294,408 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.91 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -25.49 |
| Energy contribution | -25.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
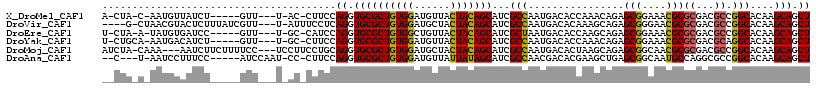
>X_DroMel_CAF1 17294303 105 - 22224390 A-CUA-C-AAUGUUAUCU-----GUU---U-AC-CUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACACCAAACAGAGCGGAAACGCGCGACGCCGGCACAAGCAGCU .-...-.-...(((.(((-----(((---(-..-..((..((((.(((((((......)))))))..))))...))....)))))))(((....)))..)))((..((....)).)). ( -32.40) >DroVir_CAF1 7060 109 - 1 ----G-CUAACGUACUCUUUAUCGUU---U-AUUUCCUCAGGUGCGCUGUGGAUGCUACUACAGCAUCGCCAAUGACACAAAGCAGAGCGGGAACGCGCGACGCCGGCACAAGCAGCU ----(-((((((..........))))---.-..........(((((((((((......))))))).((((...((........))..(((....))))))).....)))).))).... ( -29.90) >DroEre_CAF1 12263 105 - 1 U-CUA-A-UAUGUGAUCC-----GUU---U-GC-CAUCCAGGUGCGCUGUGGCUGUUACUACAGCAUCGCUAAUGACACCAAGCAGAGCGGAAACGCGCGACGCCGGCACAAGCAGCU .-...-.-..........-----(((---(-(.-...((.((((.(((((((......))))))).((((...((....))......(((....)))))))))))))..))))).... ( -32.60) >DroYak_CAF1 2480 106 - 1 U-CUGCA-AAUGACAUCU-----GUU---U-GC-CUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACACCAAACAGAGCGGAAACGCGCGACGCAGGCACAAGCAGCU .-((((.-.......(((-----(((---(-(.-..((..((((.(((((((......)))))))..))))...))...))))))))(((....))).....))))((....)).... ( -39.20) >DroMoj_CAF1 17555 111 - 1 AUCUA-CAAA---AAUCUUCUUUUCC---UCCUUCCUGCAGGUGCGCUGUGGAUGCUACUACAGCAUCGCCAAUGACACUAAGCAGAGCGGCAACGCGCGACGCCGGCACAAGCAGCU .....-....---.............---......((((..(((((((((((......))))))).((((...((........))..(((....))))))).....))))..)))).. ( -33.80) >DroAna_CAF1 10856 105 - 1 --C---U-AAUCCUUUCC-----AUCCAAU-CC-CUUCCAGGUGCGCUGUGGAUGUUAUUAUAGCAUCGCCAACGACACGAAGCUGAGCGGCAAUGCCAGGCGCCGGCACAAGCAGCU --.---.-..........-----.......-.(-(.....))...(((((.(((((((...)))))))..............((((.(((.(.......).)))))))....))))). ( -25.10) >consensus __CUA_C_AAUGUAAUCU_____GUU___U_AC_CUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACACCAAGCAGAGCGGAAACGCGCGACGCCGGCACAAGCAGCU .......................................((.((((((((((......)))))))...(((................(((....)))((...)).)))....))).)) (-25.49 = -25.27 + -0.22)


| Location | 17,294,343 – 17,294,433 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -19.42 |
| Consensus MFE | -14.68 |
| Energy contribution | -14.27 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
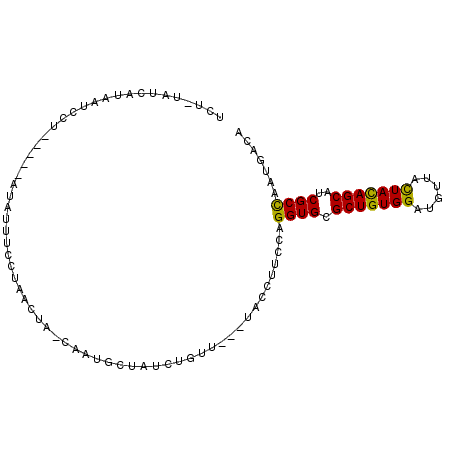
>X_DroMel_CAF1 17294343 90 - 22224390 UCU-UAUCAUAAUCCU-----AUAUUUCCUAACUA-CAAUGUUAUCUGUU---UACCUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACA ...-..((((......-----........((((..-....))))......---.........((((.(((((((......)))))))..)))).)))).. ( -18.50) >DroSec_CAF1 11350 92 - 1 UCU-UAUCAUAAUCCUUU---AUAUUUCAUAACUA-CAAUGCUAUCUGUU---UACCUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACA ...-..((((........---..............-....((.(((((..---.......)))))))(((((((......))))))).......)))).. ( -17.50) >DroSim_CAF1 2502 92 - 1 UCU-UAUCAUAAUCCUUU---AUAUUUCAUAACUA-CAAUGCUAUCUGUU---UACCUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACA ...-..((((........---..............-....((.(((((..---.......)))))))(((((((......))))))).......)))).. ( -17.50) >DroEre_CAF1 12303 90 - 1 UCU-UAUCAUUACGUU-----AUAUUUCUUAUCUA-AUAUGUGAUCCGUU---UGCCAUCCAGGUGCGCUGUGGCUGUUACUACAGCAUCGCUAAUGACA ...-..((((((....-----(((((........)-))))(((((..((.---.(((.....)))))(((((((......)))))))))))))))))).. ( -20.50) >DroYak_CAF1 2520 92 - 1 UCCUGAACAUAAUCCU-----AUCGUUCCUAUCUGCAAAUGACAUCUGUU---UGCCUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACA ....(((((((....)-----)).))))......((((((.......)))---)))......((((.(((((((......)))))))..))))....... ( -21.70) >DroAna_CAF1 10896 94 - 1 AUU-CGAC-UAAUCCUGGCAAGGAUCUCCUG-C---UAAUCCUUUCCAUCCAAUCCCUUCCAGGUGCGCUGUGGAUGUUAUUAUAGCAUCGCCAACGACA ..(-((..-......(((.((((((......-.---..)))))).)))..............((((.(((((((......)))))))..))))..))).. ( -20.80) >consensus UCU_UAUCAUAAUCCU_____AUAUUUCCUAACUA_CAAUGCUAUCUGUU___UACCUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACA ..............................................................((((.(((((((......)))))))..))))....... (-14.68 = -14.27 + -0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:46 2006