
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,290,184 – 17,290,343 |
| Length | 159 |
| Max. P | 0.938393 |

| Location | 17,290,184 – 17,290,303 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -40.44 |
| Consensus MFE | -34.26 |
| Energy contribution | -35.04 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.03 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.932981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17290184 119 + 22224390 AAGCAAC-UGGCUCGCUGUGGAUAGAUAAUUCACAGACGAUGCCGAAUAAUAUGCCAUCCAAGUACAUGUACAUUUGGGUUUCCCGGGGAACCCGUAAAUAUGUGUGAUAUAUAGGUAAA .......-(((((((((((((((.....)))))))).))).)))).......((((.......((((((((....((((((((....))))))))....)))))))).......)))).. ( -40.74) >DroSim_CAF1 9014 119 + 1 AAGCAACUUGGCUCGCUGUGGAUAGAUAAUUCACAGACGAUGCCGAA-AAUAUGCCAUCCAAGUACAUGUACAUUUGGGUUUCCCGGGGAACCCGUAAAUAUGUGUGAUAUAUAGGUAAA .......((((((((((((((((.....)))))))).))).))))).-....((((.......((((((((....((((((((....))))))))....)))))))).......)))).. ( -42.04) >DroYak_CAF1 17628 120 + 1 AAGCAACUGGCUUCCCUGUGGAUAGAUAAUUCACAAUCGAAGCCGAAUAAUAUGCCAUCCAAGUACAUGUACAUGUGGGUUUCCCGGGGAACCCGUAAAUAUGUGUGAUAUAUAGGUAAA .......(((((((..(((((((.....)))))))...))))))).......((((.......((((((((..((((((((((....))))))))))..)))))))).......)))).. ( -38.54) >consensus AAGCAACUUGGCUCGCUGUGGAUAGAUAAUUCACAGACGAUGCCGAAUAAUAUGCCAUCCAAGUACAUGUACAUUUGGGUUUCCCGGGGAACCCGUAAAUAUGUGUGAUAUAUAGGUAAA .........((((((((((((((.....)))))))).))).)))........((((.......((((((((....((((((((....))))))))....)))))))).......)))).. (-34.26 = -35.04 + 0.78)

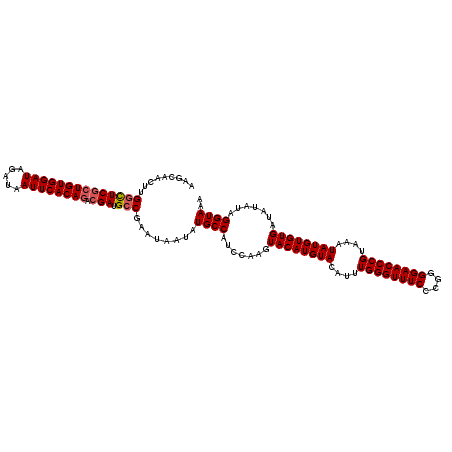

| Location | 17,290,223 – 17,290,343 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -32.47 |
| Energy contribution | -32.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17290223 120 + 22224390 UGCCGAAUAAUAUGCCAUCCAAGUACAUGUACAUUUGGGUUUCCCGGGGAACCCGUAAAUAUGUGUGAUAUAUAGGUAAAUAUAUUAUUUUCAUAUAUGUCGAUGAGUGCGUUAACGCGU .............((((((.....(((((((....((((((((....))))))))....)))))))((((((((...(((((...)))))...)))))))))))).))(((....))).. ( -32.70) >DroSim_CAF1 9054 119 + 1 UGCCGAA-AAUAUGCCAUCCAAGUACAUGUACAUUUGGGUUUCCCGGGGAACCCGUAAAUAUGUGUGAUAUAUAGGUAAAUAUAUUAUUUUCAUAUAUGUCGAUGAGUGCGUUAACGCGU .......-.....((((((.....(((((((....((((((((....))))))))....)))))))((((((((...(((((...)))))...)))))))))))).))(((....))).. ( -32.70) >DroYak_CAF1 17668 120 + 1 AGCCGAAUAAUAUGCCAUCCAAGUACAUGUACAUGUGGGUUUCCCGGGGAACCCGUAAAUAUGUGUGAUAUAUAGGUAAAUAUAUUAUUUUCAUAUAUGUCGAUGAGUGCGUUAACGCGU .............((((((.....(((((((..((((((((((....))))))))))..)))))))((((((((...(((((...)))))...)))))))))))).))(((....))).. ( -32.90) >consensus UGCCGAAUAAUAUGCCAUCCAAGUACAUGUACAUUUGGGUUUCCCGGGGAACCCGUAAAUAUGUGUGAUAUAUAGGUAAAUAUAUUAUUUUCAUAUAUGUCGAUGAGUGCGUUAACGCGU .............((((((.....(((((((....((((((((....))))))))....)))))))((((((((...(((((...)))))...)))))))))))).))(((....))).. (-32.47 = -32.47 + -0.00)

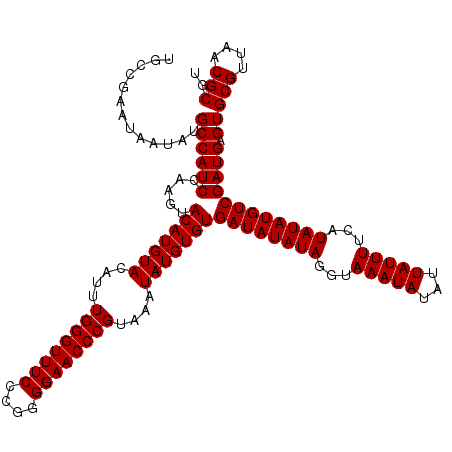

| Location | 17,290,223 – 17,290,343 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -24.10 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17290223 120 - 22224390 ACGCGUUAACGCACUCAUCGACAUAUAUGAAAAUAAUAUAUUUACCUAUAUAUCACACAUAUUUACGGGUUCCCCGGGAAACCCAAAUGUACAUGUACUUGGAUGGCAUAUUAUUCGGCA ..(((....)))..(((((.(..((((((......((((((......))))))...((((......(((...)))((....))...)))).))))))..).))))).............. ( -24.10) >DroSim_CAF1 9054 119 - 1 ACGCGUUAACGCACUCAUCGACAUAUAUGAAAAUAAUAUAUUUACCUAUAUAUCACACAUAUUUACGGGUUCCCCGGGAAACCCAAAUGUACAUGUACUUGGAUGGCAUAUU-UUCGGCA ..(((....)))..(((((.(..((((((......((((((......))))))...((((......(((...)))((....))...)))).))))))..).)))))......-....... ( -24.10) >DroYak_CAF1 17668 120 - 1 ACGCGUUAACGCACUCAUCGACAUAUAUGAAAAUAAUAUAUUUACCUAUAUAUCACACAUAUUUACGGGUUCCCCGGGAAACCCACAUGUACAUGUACUUGGAUGGCAUAUUAUUCGGCU ..(((....)))..(((((.(..((((((......((((((......))))))...((((......(((...)))((....))...)))).))))))..).))))).............. ( -24.10) >consensus ACGCGUUAACGCACUCAUCGACAUAUAUGAAAAUAAUAUAUUUACCUAUAUAUCACACAUAUUUACGGGUUCCCCGGGAAACCCAAAUGUACAUGUACUUGGAUGGCAUAUUAUUCGGCA ..(((....)))..(((((.(..((((((......((((((......))))))...((((......(((...)))((....))...)))).))))))..).))))).............. (-24.10 = -24.10 + -0.00)

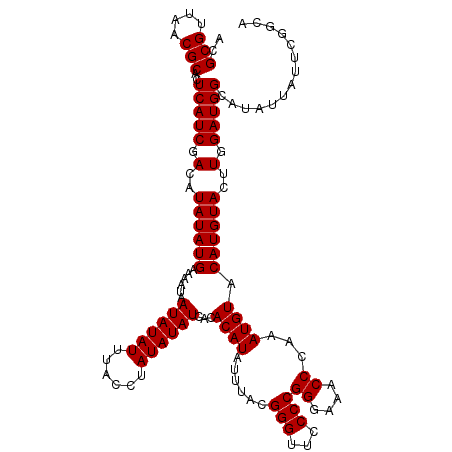

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:44 2006