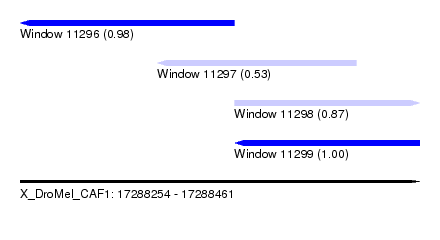
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,288,254 – 17,288,461 |
| Length | 207 |
| Max. P | 0.998617 |

| Location | 17,288,254 – 17,288,365 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -35.11 |
| Energy contribution | -34.67 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17288254 111 - 22224390 GCCAGACAUUAUGCAAUAUUGGGGCCCUAAGGCGAUUGGGACCGGUACCCGAAAUAGCCACCGAAAUACGCUGCCAUUCAAAUGCAGUGAUGCCCCAAUAGUUUGAAAU---------AU ..(((((.........(((((((((.....(((..(((((.......)))))....))).........((((((.........))))))..))))))))))))))....---------.. ( -35.20) >DroSim_CAF1 7126 111 - 1 GCCAGACAUUAUGCAAUAUUGGGGUCCUAAGGCGAUUGGGACCGGUACCCGAAAUAGCCACCGAAAUACGCUGCCAUUCAAAUGCAGUGAUGCCCCAAUAGUUUGAAAU---------AU ..(((((.........(((((((((.....(((..(((((.......)))))....))).........((((((.........))))))..))))))))))))))....---------.. ( -33.30) >DroYak_CAF1 8694 120 - 1 GCCAGACAUUAUGCAAUAUUGGGGCCCUAAGGCGAUUGGGACCGGUACCCAAAAUAGCCACCGAAAUACGCUGCCAUUCAAAUGCAGUGAUGCCCCAAUAGUUUGAAAUCGAAUAGGUGU (((.............(((((((((.....(((..(((((.......)))))....))).........((((((.........))))))..)))))))))(((((....))))).))).. ( -39.10) >consensus GCCAGACAUUAUGCAAUAUUGGGGCCCUAAGGCGAUUGGGACCGGUACCCGAAAUAGCCACCGAAAUACGCUGCCAUUCAAAUGCAGUGAUGCCCCAAUAGUUUGAAAU_________AU ..(((((.........(((((((((.....(((..(((((.......)))))....))).........((((((.........))))))..))))))))))))))............... (-35.11 = -34.67 + -0.44)


| Location | 17,288,325 – 17,288,428 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.09 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -30.87 |
| Energy contribution | -31.10 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17288325 103 - 22224390 UUGGUCAUC-----------------GGCCAUUGGCCAUUGGCCAUCGGACACCGGCCACCGGCUGAUAAAUGAGUCUGGGCCAGACAUUAUGCAAUAUUGGGGCCCUAAGGCGAUUGGG (..((((((-----------------((((..(((((..((.((...)).))..)))))..)))))))......(((((((((...((.(((...))).)).)))))..)))))))..). ( -40.40) >DroSim_CAF1 7197 103 - 1 UUGGUCAUC-----------------GGCCAUUGGCCAUUGGCCAUCGGACACCGGCCACCGGCUGACAAAUGAGUCUGGGCCAGACAUUAUGCAAUAUUGGGGUCCUAAGGCGAUUGGG (..(((.((-----------------((((..(((((..((.((...)).))..)))))..)))))).......(((((((((...((.(((...))).)).)))))..)))))))..). ( -36.40) >DroYak_CAF1 8774 120 - 1 UUGGUCAUCAUCGGCCACCAGCCACCAGCCACCGGCCAUCGGGCAUUGGCCAUUGGCCAUCGGCUGAUAAAUGAGUCUGGGCCAGACAUUAUGCAAUAUUGGGGCCCUAAGGCGAUUGGG (..(((.((((.(((.....)))..(((((...(((((...(((....)))..)))))...)))))....))))(((((((((...((.(((...))).)).)))))..)))))))..). ( -48.30) >consensus UUGGUCAUC_________________GGCCAUUGGCCAUUGGCCAUCGGACACCGGCCACCGGCUGAUAAAUGAGUCUGGGCCAGACAUUAUGCAAUAUUGGGGCCCUAAGGCGAUUGGG (..(((....................((((..(((((..((.........))..)))))..)))).........(((((((((...((.(((...))).)).)))))..)))))))..). (-30.87 = -31.10 + 0.23)

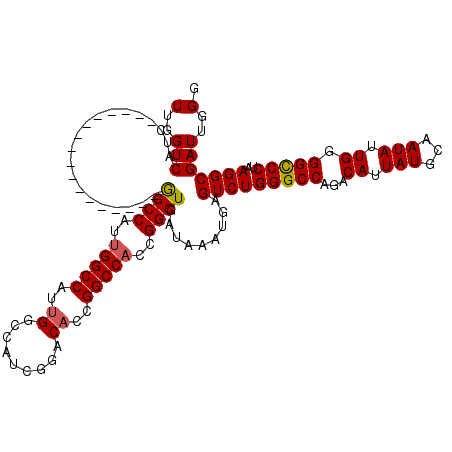
| Location | 17,288,365 – 17,288,461 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.38 |
| Mean single sequence MFE | -41.40 |
| Consensus MFE | -26.92 |
| Energy contribution | -25.59 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17288365 96 + 22224390 CCAGACUCAUUUAUCAGCCGGUGGCCGGUGUCCGAUGGCCAAUGGCCAAUGGCC-----------------GAUGACCAAAGGCCAGCGGA-------GGAGAGUCAGCCGAACUGCAAU ...(((((.....((..(((.(((((..((..((.((((((........)))))-----------------).))..))..))))).))).-------.))))))).((......))... ( -39.30) >DroSim_CAF1 7237 96 + 1 CCAGACUCAUUUGUCAGCCGGUGGCCGGUGUCCGAUGGCCAAUGGCCAAUGGCC-----------------GAUGACCAAAGGCCAGCGGA-------GGAGAGCCAGCCGAACUGCAAU ...(((......))).(((((((((...(.((((.(((((...((((...))))-----------------(.....)...))))).))))-------.)...)))).)))....))... ( -36.80) >DroYak_CAF1 8814 120 + 1 CCAGACUCAUUUAUCAGCCGAUGGCCAAUGGCCAAUGCCCGAUGGCCGGUGGCUGGUGGCUGGUGGCCGAUGAUGACCAAAGGCCAGCGGAGAGGAGAGAAGAGCCAGUCGAACUGCAAU .....(((..((((((((((.((((((..(((....)))...)))))).))))))))))(((.(((((..((.....))..))))).))).))).........((.((.....))))... ( -48.10) >consensus CCAGACUCAUUUAUCAGCCGGUGGCCGGUGUCCGAUGGCCAAUGGCCAAUGGCC_________________GAUGACCAAAGGCCAGCGGA_______GGAGAGCCAGCCGAACUGCAAU .(((............((((.((((((..(((....)))...)))))).))))............................(((..((...............))..)))...))).... (-26.92 = -25.59 + -1.32)

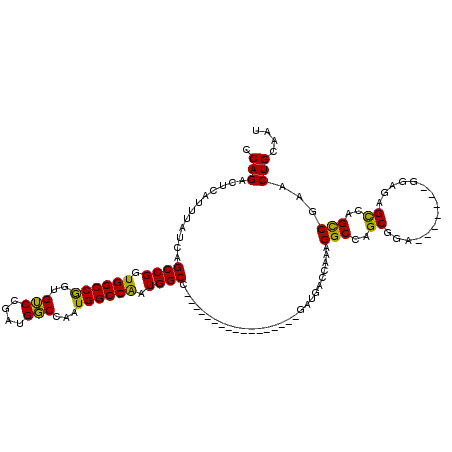
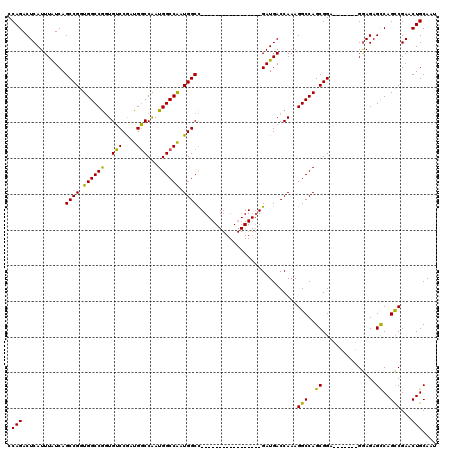
| Location | 17,288,365 – 17,288,461 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.38 |
| Mean single sequence MFE | -42.67 |
| Consensus MFE | -31.65 |
| Energy contribution | -31.43 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.74 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17288365 96 - 22224390 AUUGCAGUUCGGCUGACUCUCC-------UCCGCUGGCCUUUGGUCAUC-----------------GGCCAUUGGCCAUUGGCCAUCGGACACCGGCCACCGGCUGAUAAAUGAGUCUGG ...((......)).(((((...-------((((.(((((..((((((..-----------------......))))))..))))).))))...(((((...)))))......)))))... ( -39.50) >DroSim_CAF1 7237 96 - 1 AUUGCAGUUCGGCUGGCUCUCC-------UCCGCUGGCCUUUGGUCAUC-----------------GGCCAUUGGCCAUUGGCCAUCGGACACCGGCCACCGGCUGACAAAUGAGUCUGG .(((((((.(((.(((((....-------((((.(((((..((((((..-----------------......))))))..))))).))))....)))))))))))).))).......... ( -44.90) >DroYak_CAF1 8814 120 - 1 AUUGCAGUUCGACUGGCUCUUCUCUCCUCUCCGCUGGCCUUUGGUCAUCAUCGGCCACCAGCCACCAGCCACCGGCCAUCGGGCAUUGGCCAUUGGCCAUCGGCUGAUAAAUGAGUCUGG ............(.(((((.............(((((....(((((......))))))))))...(((((...(((((...(((....)))..)))))...)))))......))))).). ( -43.60) >consensus AUUGCAGUUCGGCUGGCUCUCC_______UCCGCUGGCCUUUGGUCAUC_________________GGCCAUUGGCCAUUGGCCAUCGGACACCGGCCACCGGCUGAUAAAUGAGUCUGG .(((((((.(((.(((((............(((.(((((..(((((....................)))))..))))).)))............)))))))))))).))).......... (-31.65 = -31.43 + -0.22)

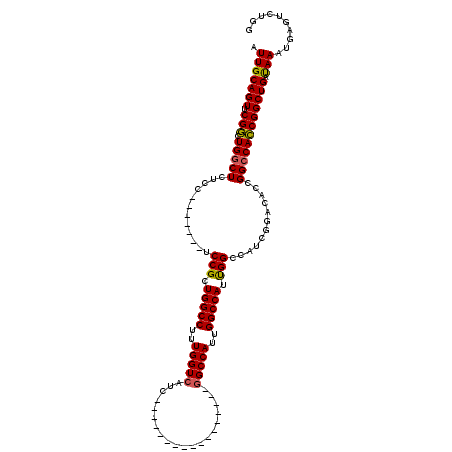
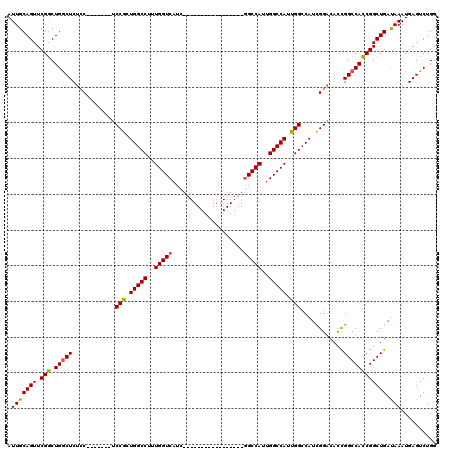
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:41 2006