
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,286,477 – 17,286,634 |
| Length | 157 |
| Max. P | 0.526555 |

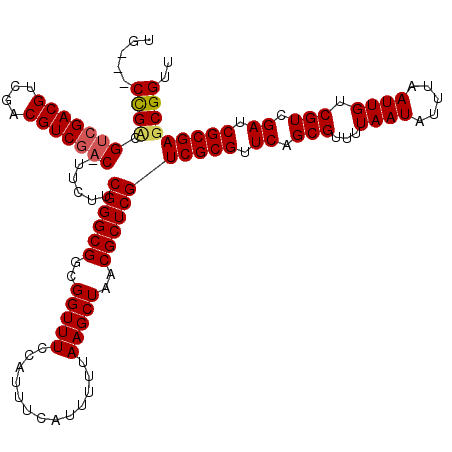
| Location | 17,286,477 – 17,286,596 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.10 |
| Mean single sequence MFE | -39.31 |
| Consensus MFE | -34.02 |
| Energy contribution | -34.69 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17286477 119 - 22224390 AGUCGCUGCCGUCGACGUCGACGUCCAC-UUAUUCGGGCGGCGGUUUCCAUUUCAUUUUUAAGCUAACGCUCGUCGCGUUCAGCGUUUUAAUAUUUAAUUGUCGUCGAUCGCGAGCGGUU ..(((((((.(((((((.((((((((..-......)))))......................((((((((.....))))).)))..............))).))))))).)).))))).. ( -39.60) >DroSim_CAF1 5404 117 - 1 UG---CCGUCGUCGACGUCGACGUCGACGUUCUUCGGGCGGCGGUUUCCAUUUCAUUUUUAAGCUAACGCUCGUCGCGUUCAGCGUUUUAAUAUUUAAUUGUCGUCGAUCGCGAGCGGUU .(---((((((((((((....)))))))).....((((((..(((((.............)))))..))))))(((((.((.(((...((((.....)))).))).)).)))))))))). ( -42.72) >DroYak_CAF1 6721 116 - 1 UG---CCGACGCCGACGUCGACGUCGAC-UUCUUCGGGCGGCGGUUUCCAUUUCAUUUUUAAGCUAACGCUCGUCGCGUUCAGCGUUUUAAUAUUUAAUUGUCGUCGAUCGCGAGCGGUU ..---....(((((.((((((((.(((.-.....((((((..(((((.............)))))..)))))).(((.....))).............))).)))))).)))).)))... ( -35.62) >consensus UG___CCGACGUCGACGUCGACGUCGAC_UUCUUCGGGCGGCGGUUUCCAUUUCAUUUUUAAGCUAACGCUCGUCGCGUUCAGCGUUUUAAUAUUUAAUUGUCGUCGAUCGCGAGCGGUU .....((((.(((((((....)))))))......((((((..(((((.............)))))..))))))(((((.((.(((...((((.....)))).))).)).))))))))).. (-34.02 = -34.69 + 0.67)


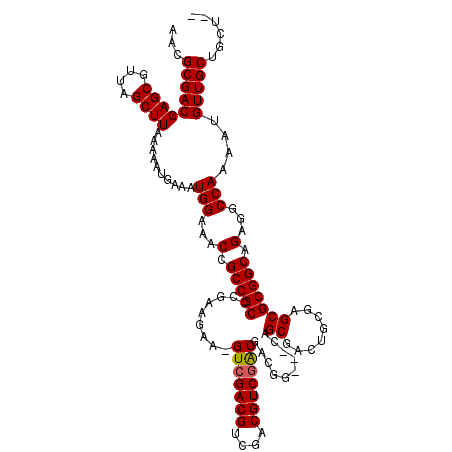
| Location | 17,286,517 – 17,286,634 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -36.52 |
| Energy contribution | -36.63 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17286517 117 + 22224390 AACGCGACGAGCGUUAGCUUAAAAAUGAAAUGGAAACCGCCGCCCGAAUAA-GUGGACGUCGACGUCGACGGCAGCGACUGCGACGGCGAGCGCGGCAGAGGCCAAAAUGUUGCUGCU-- ...((((((.(((((................(....)(((((..((....(-((.(.(((((....)))))....).))).)).))))))))))(((....)))....))))))....-- ( -41.00) >DroSim_CAF1 5444 115 + 1 AACGCGACGAGCGUUAGCUUAAAAAUGAAAUGGAAACCGCCGCCCGAAGAACGUCGACGUCGACGUCGACGACGG---CAGCGACUGCGAGCGCGGCAGAGGCCAAAAUGUUGCUGCU-- ...((((((.(((((.((.............(....)(((.(((.......((((((((....))))))))..))---).)))...)).)))))(((....)))....))))))....-- ( -43.30) >DroYak_CAF1 6761 116 + 1 AACGCGACGAGCGUUAGCUUAAAAAUGAAAUGGAAACCGCCGCCCGAAGAA-GUCGACGUCGACGUCGGCGUCGG---CAGCGACUGCGAGCGCGGCUGAGGCCAAAAUGUUGCUGCGGA ...(((((((((....))))..........(((...(.((((((((....(-((((..(((((((....))))))---)..))))).)).).))))).)...)))....)))))...... ( -46.20) >consensus AACGCGACGAGCGUUAGCUUAAAAAUGAAAUGGAAACCGCCGCCCGAAGAA_GUCGACGUCGACGUCGACGACGG___CAGCGACUGCGAGCGCGGCAGAGGCCAAAAUGUUGCUGCU__ ...(((((((((....))))..........(((...(.(((((.........(((((((....)))))))..........((........))))))).)...)))....)))))...... (-36.52 = -36.63 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:37 2006