
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,260,985 – 17,261,216 |
| Length | 231 |
| Max. P | 0.934715 |

| Location | 17,260,985 – 17,261,097 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -20.06 |
| Energy contribution | -21.98 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17260985 112 - 22224390 GCUCGUUUUGCUUUUGACAGUUCAUGAUAGAUUGAAUUGCAGA-AGCUGUUGAUAAAUUGGACAGCUGAUACAAGGCCAAAGUUUGCUCCUA-------UGGCGAAUUGGUUGCAAGAGG ...(.((((((.((((.(((((((........)))))))))))-(((((((.(.....).))))))).......(((((..(((((((....-------.)))))))))))))))))).) ( -34.30) >DroSec_CAF1 106514 109 - 1 GCUCGUUUUGCGUUUGACAGUUCA---UAGAUUGAAUUGCAGA-AGCUGUUGAUAAAUUGGACAGCUGAUGCGAGACCAAAGUUUGCUCCUA-------UGGCGAAUUGGUUGCAAGAGG .(((((((((((((((.(((((((---.....)))))))))..-(((((((.(.....).)))))))))))))))))...((((((((....-------.))))))))........))). ( -36.70) >DroSim_CAF1 110085 112 - 1 GCUCGUUUUGCUUUUGACAGUCCAUGAUAGAUUGAAUUGCAGA-AGCUGUUGAUAAAUUGGACAGCUGAUGCGAGACCAAAGUUAGCUCCUA-------UGGCGAAUUGGUUGCAAGAGG ...(.((((((......(((((.......)))))..(((((..-(((((((.(.....).)))))))..)))))((((((.....(((....-------.)))...)))))))))))).) ( -33.50) >DroEre_CAF1 112025 99 - 1 GCUCGUUGUGCCUUCGACAGCUCAUGAUAGAUUGAAUUGGAGA-AGCUGUUGAUGAAUUGGGCAGCUG--GCA-----GAAGUUUGCUUCUA-------UGGCGAAUUUGAUGU------ ..((((..((((.((..(((.(((........))).)))..))-(((((((.(.....).))))))))--))(-----((((....))))))-------..)))).........------ ( -30.30) >DroYak_CAF1 116472 119 - 1 GCUCGUUGUGCUUUUGACAGUUCAUGAUAGAUGGAAUUGAAGAAAGCUGUUGAUAAAUUGGACAGCUGAGGCAAGAGCAAAGUUGGCUUCCAUCAAACAUGUCAAAUUUGUUGCAA-AGG ((((....((((((...((((((..........)))))).....(((((((.(.....).))))))))))))).))))(((.(((((.............))))).))).......-... ( -28.82) >consensus GCUCGUUUUGCUUUUGACAGUUCAUGAUAGAUUGAAUUGCAGA_AGCUGUUGAUAAAUUGGACAGCUGAUGCAAGACCAAAGUUUGCUCCUA_______UGGCGAAUUGGUUGCAAGAGG ((..(((((((......(((((((........))))))).....(((((((.(.....).)))))))...)))))))...((((((((............))))))))....))...... (-20.06 = -21.98 + 1.92)

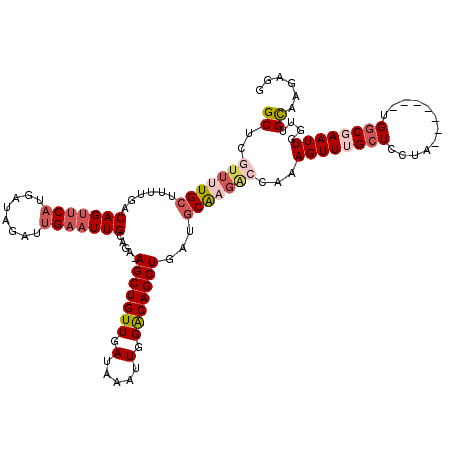
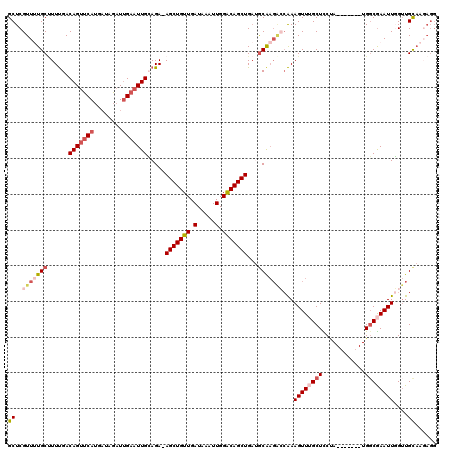
| Location | 17,261,097 – 17,261,216 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.97 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -9.68 |
| Energy contribution | -9.84 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17261097 119 + 22224390 UUUUCUAUUUUCACACUACAAUCAAGUUGACUUAUUGCACUUUGCACAAGUCAAUGGAACGCAUAAGUUCGAUUGCACAUAAGAUAACAUCCGCAUGGAAUUUAAAAAA-UUCGGAAUAA .....((((((.......(((((..((((((((..(((.....))).)))))))).((((......))))))))).....))))))...((((.((............)-).)))).... ( -23.00) >DroSec_CAF1 106623 119 + 1 UCUUCUAUUUUCACACUACGAGUAAGUUGACUUAUUGCACUUUGCUCAAGUCAGUGGAACGCAUAAGUUGGCUUGCACAUAAGAUAACAUCCGCAUGGAAUUUCAAUAA-UUCCGAAUAA .........(((.........((((((..((((((.((..((..((......))..))..))))))))..))))))....................((((((.....))-)))))))... ( -29.40) >DroSim_CAF1 110197 119 + 1 UCUUCUAUUUUCACACUACGAGCAAGUUGACUUAUUGCACUUUGCACAAGUCAGUGGAACGCAUAAGUUGGCUUGCACAUAAGAUAACAUCCGCAUGGAAUUUCAAUAA-UUCCGAAUAA .........(((.........((((((..((((((.((..((..(........)..))..))))))))..))))))....................((((((.....))-)))))))... ( -30.50) >DroEre_CAF1 112124 96 + 1 UCUUUUAUACUCACACCACCAGCAAGUUGACCUAUUGCACUUUGCACAAGUCAGCGGAGCACAGAAGA---CACACAUC------------CGUGUGAAAUUUCAAUACAU--------- ..........((((((.........((((((....(((.....)))...))))))(((..........---......))------------))))))).............--------- ( -19.69) >DroYak_CAF1 116591 101 + 1 UCUUCUAUCUUCACGCCACAAGGAGGUUGACUCAUUGCACUUAGCACCAGUCAAUGGAACGCACAAGUUGGCCUGCAUGCAGGAUAAGAUAAGAGUGAAGA------------------- .......((((((((((........(((((((...(((.....)))..)))))))..(((......))))))(((....)))............)))))))------------------- ( -26.60) >consensus UCUUCUAUUUUCACACUACAAGCAAGUUGACUUAUUGCACUUUGCACAAGUCAGUGGAACGCAUAAGUUGGCUUGCACAUAAGAUAACAUCCGCAUGGAAUUUCAAUAA_UUC_GAAUAA ..((((((.................(((((((...(((.....)))..))))))).....(((..........)))..................)))))).................... ( -9.68 = -9.84 + 0.16)

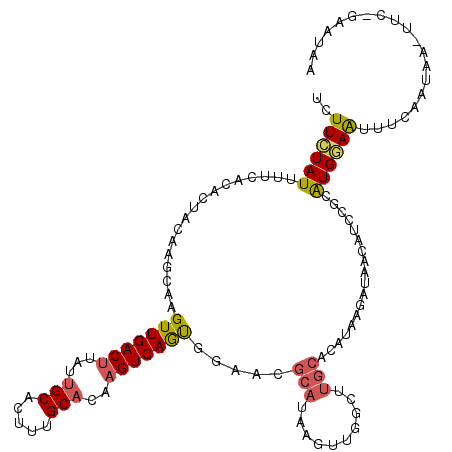
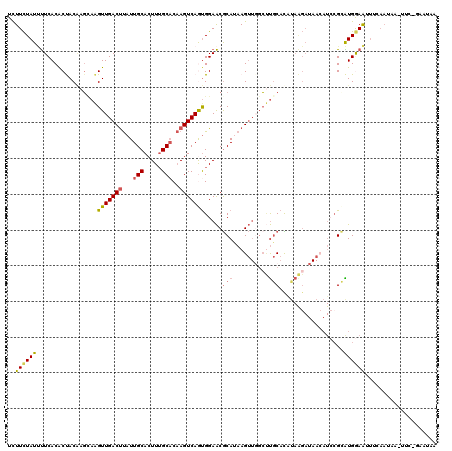
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:28 2006