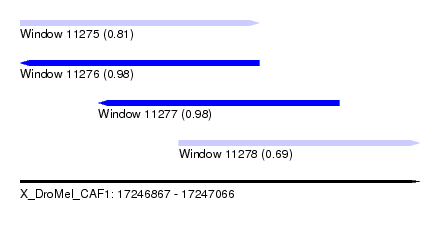
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,246,867 – 17,247,066 |
| Length | 199 |
| Max. P | 0.983944 |

| Location | 17,246,867 – 17,246,986 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.98 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -32.28 |
| Energy contribution | -32.24 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17246867 119 + 22224390 -GGGUAGGAGAAUUGCCCCCUGCUAAGUGACAAUUAUUCGUGCUAGUUGUCGGUUUGGUUCUCCAAAGUAUUGGUUUCGUUACGCGAUUGGUUAGCGUUAAGUGGACGCCAAUCGCAUUU -.....(((((((((..((((....)).((((((((.......))))))))))..)))))))))...(((.((....)).)))(((((((((..((.....))....))))))))).... ( -36.20) >DroSec_CAF1 92545 119 + 1 -GGGUGGUAGGAUUGCGCCCUGCUAAGUGACAAUUAUUCGUGCUAGUUGUCGGUUUGGUUCUCCAAAGUAUUGGUUUCGUUACGCGAUUGGUUAGCGUUAAGUGGACGCCAAUCGCAUUU -(((((.........))))).(((((..((((((((.......))))))))(.(((((....))))).).)))))........(((((((((..((.....))....))))))))).... ( -35.90) >DroSim_CAF1 94978 119 + 1 -GGGUGGUAGGAUUGCCCCCUGCUAAGUGACAAUUAUUCGUGCUAGUUGUCGGUUUGGUUCUCCAAAGUAUUGGUUUCGUUACGCGAUUGGUUAGCGUUAAGUGGACGCCAAUCGCGUUU -(((.((((....))))))).(((((..((((((((.......))))))))(.(((((....))))).).)))))......(((((((((((..((.....))....))))))))))).. ( -39.90) >DroEre_CAF1 98865 119 + 1 GUGGAAGUGGGAUUGCC-CCUGCUAAGUGACAAUUAUUCGUGCUAGUUGUCGGUUUGGUUCUCCAAAGUAUUGGUUACGUUACGCGAUUGGCUGGCGUUAAGUGGACGCCAAUCGCAUUU .((((((..((......-))..))((.(((((((((.......))))))))).))......))))..(((.((....)).)))((((((((((.((.....)).)..))))))))).... ( -37.40) >DroYak_CAF1 102029 120 + 1 NNNGAAGGGGGAUUGCCCCCUGCUAAGUGACAAUUAUUCGUGCUAGUUGUCGGUUUGGUUCUCCAAAGUAUUGGUUACGUUACGCGAUUGGUUAGCGUUAAGUGGACGCCAAUCGCAUUU .....((((((....))))))(((((..((((((((.......))))))))(.(((((....))))).).)))))........(((((((((..((.....))....))))))))).... ( -40.70) >consensus _GGGUAGUAGGAUUGCCCCCUGCUAAGUGACAAUUAUUCGUGCUAGUUGUCGGUUUGGUUCUCCAAAGUAUUGGUUUCGUUACGCGAUUGGUUAGCGUUAAGUGGACGCCAAUCGCAUUU .....((((((.......))))))...(((((((((.......))))))))).(((((....)))))(((.((....)).)))(((((((((..((.....))....))))))))).... (-32.28 = -32.24 + -0.04)

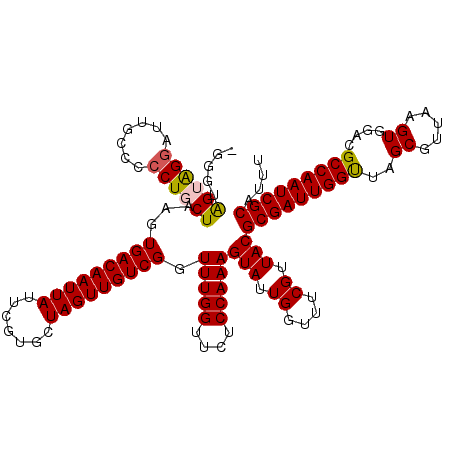

| Location | 17,246,867 – 17,246,986 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.98 |
| Mean single sequence MFE | -28.99 |
| Consensus MFE | -24.16 |
| Energy contribution | -24.00 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17246867 119 - 22224390 AAAUGCGAUUGGCGUCCACUUAACGCUAACCAAUCGCGUAACGAAACCAAUACUUUGGAGAACCAAACCGACAACUAGCACGAAUAAUUGUCACUUAGCAGGGGGCAAUUCUCCUACCC- ..(((((((((((((.......))))....))))))))).................((((((.....(((((((.((.......)).))))).((.....))))....)))))).....- ( -27.80) >DroSec_CAF1 92545 119 - 1 AAAUGCGAUUGGCGUCCACUUAACGCUAACCAAUCGCGUAACGAAACCAAUACUUUGGAGAACCAAACCGACAACUAGCACGAAUAAUUGUCACUUAGCAGGGCGCAAUCCUACCACCC- ......(((((.(((((.....((((.........)))).......((((....))))...........(((((.((.......)).)))))........)))))))))).........- ( -25.10) >DroSim_CAF1 94978 119 - 1 AAACGCGAUUGGCGUCCACUUAACGCUAACCAAUCGCGUAACGAAACCAAUACUUUGGAGAACCAAACCGACAACUAGCACGAAUAAUUGUCACUUAGCAGGGGGCAAUCCUACCACCC- ..(((((((((((((.......))))....))))))))).......((((....))))...........(((((.((.......)).)))))........(((((........)).)))- ( -29.00) >DroEre_CAF1 98865 119 - 1 AAAUGCGAUUGGCGUCCACUUAACGCCAGCCAAUCGCGUAACGUAACCAAUACUUUGGAGAACCAAACCGACAACUAGCACGAAUAAUUGUCACUUAGCAGG-GGCAAUCCCACUUCCAC ..(((((((((((...............)))))))))))................(((((.........(((((.((.......)).)))))........((-(.....)))..))))). ( -30.46) >DroYak_CAF1 102029 120 - 1 AAAUGCGAUUGGCGUCCACUUAACGCUAACCAAUCGCGUAACGUAACCAAUACUUUGGAGAACCAAACCGACAACUAGCACGAAUAAUUGUCACUUAGCAGGGGGCAAUCCCCCUUCNNN ..(((((((((((((.......))))....)))))))))...((..((((....))))...........(((((.((.......)).))))).....))((((((....))))))..... ( -32.60) >consensus AAAUGCGAUUGGCGUCCACUUAACGCUAACCAAUCGCGUAACGAAACCAAUACUUUGGAGAACCAAACCGACAACUAGCACGAAUAAUUGUCACUUAGCAGGGGGCAAUCCUACUACCC_ ..(((((((((((((.......))))....))))))))).......((.....(((((....)))))..(((((.((.......)).)))))........)).................. (-24.16 = -24.00 + -0.16)

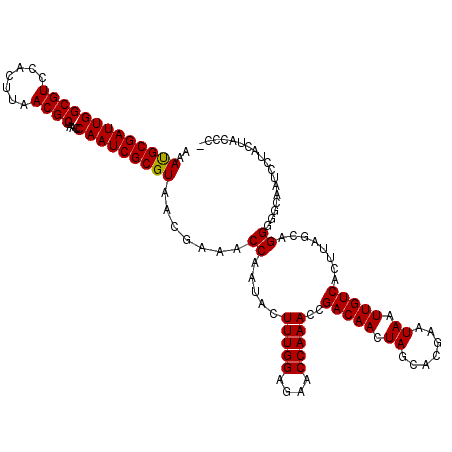

| Location | 17,246,906 – 17,247,026 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.04 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17246906 120 - 22224390 GUUAAUAUAAUUAGGCGGGAAUUACUUCGCUCUGAUCACCAAAUGCGAUUGGCGUCCACUUAACGCUAACCAAUCGCGUAACGAAACCAAUACUUUGGAGAACCAAACCGACAACUAGCA ((((......(((((((((......))))).))))((.....(((((((((((((.......))))....))))))))).......((((....)))).))..............)))). ( -27.10) >DroSec_CAF1 92584 120 - 1 GUUAAUAUAAUUAGGCGGGAAUUACUCAGCUCUGAUCACCAAAUGCGAUUGGCGUCCACUUAACGCUAACCAAUCGCGUAACGAAACCAAUACUUUGGAGAACCAAACCGACAACUAGCA ((((......(((((((((.....))).)).))))((.....(((((((((((((.......))))....))))))))).......((((....)))).))..............)))). ( -25.30) >DroSim_CAF1 95017 120 - 1 GUUAAUAUAAUUAGGCGGGAAUUACUCCGCUCUGAUCACCAAACGCGAUUGGCGUCCACUUAACGCUAACCAAUCGCGUAACGAAACCAAUACUUUGGAGAACCAAACCGACAACUAGCA ((((......(((((((((......))))).))))((.....(((((((((((((.......))))....))))))))).......((((....)))).))..............)))). ( -31.70) >DroEre_CAF1 98904 118 - 1 GUUAGCAUAAUUAGGCGGAAAUUA--CUGCUCCGAUCACCAAAUGCGAUUGGCGUCCACUUAACGCCAGCCAAUCGCGUAACGUAACCAAUACUUUGGAGAACCAAACCGACAACUAGCA (((((........(((((......--)))))....((.(((((((((((((((...............)))))))))))...(((.....))).)))).)).............))))). ( -31.16) >DroYak_CAF1 102069 120 - 1 GUCAACAUAAUUAGGCGGAAAUUACGCUGCUCCGAUCAGCAAAUGCGAUUGGCGUCCACUUAACGCUAACCAAUCGCGUAACGUAACCAAUACUUUGGAGAACCAAACCGACAACUAGCA (((..........((((.......)))).((((((.......(((((((((((((.......))))....)))))))))...(((.....))).)))))).........)))........ ( -30.10) >consensus GUUAAUAUAAUUAGGCGGGAAUUACUCCGCUCUGAUCACCAAAUGCGAUUGGCGUCCACUUAACGCUAACCAAUCGCGUAACGAAACCAAUACUUUGGAGAACCAAACCGACAACUAGCA (((.......((((((((........)))).))))((.....(((((((((((((.......))))....))))))))).......((((....)))).))........)))........ (-25.08 = -25.04 + -0.04)


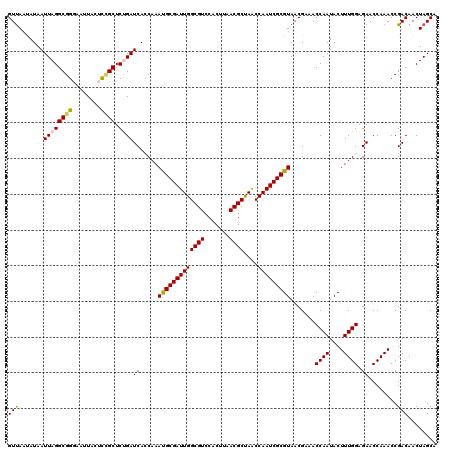
| Location | 17,246,946 – 17,247,066 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.42 |
| Mean single sequence MFE | -32.96 |
| Consensus MFE | -22.90 |
| Energy contribution | -23.06 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17246946 120 + 22224390 UACGCGAUUGGUUAGCGUUAAGUGGACGCCAAUCGCAUUUGGUGAUCAGAGCGAAGUAAUUCCCGCCUAAUUAUAUUAACUUAAUGGUUUUAAAAGCAACUCAUGUCGAUUAUUAAAAAU ...(((((((((..((.....))....))))))))).((((((((((((.(((((....)))..))))...............((((((........))).)))...))))))))))... ( -28.20) >DroSec_CAF1 92624 120 + 1 UACGCGAUUGGUUAGCGUUAAGUGGACGCCAAUCGCAUUUGGUGAUCAGAGCUGAGUAAUUCCCGCCUAAUUAUAUUAACUUAAUGGCUUUAAAAGGAACUCAAGCCGACUAUUAAGAAU ...(((((((((..((.....))....))))))))).(((((((.((.(.(((((((...(((.(((..((((........))))))).......))))))).)))))).)))))))... ( -29.60) >DroSim_CAF1 95057 120 + 1 UACGCGAUUGGUUAGCGUUAAGUGGACGCCAAUCGCGUUUGGUGAUCAGAGCGGAGUAAUUCCCGCCUAAUUAUAUUAACUUAAUGGUUUUAAAAGGAACUCAAGCCGACUAUUAAGAAU .(((((((((((..((.....))....)))))))))))...(((((.((.((((........)))))).))))).....((((((((((......((........))))))))))))... ( -37.20) >DroEre_CAF1 98944 116 + 1 UACGCGAUUGGCUGGCGUUAAGUGGACGCCAAUCGCAUUUGGUGAUCGGAGCAG--UAAUUUCCGCCUAAUUAUGCUAACUUAAUGCCCUAG--AACCACUCAGGCCGAUUAUCAAAAAU ...((((((((((.((.....)).)..))))))))).((((((((((((((((.--(((((.......)))))))))..........(((.(--(.....)))))))))))))))))... ( -39.10) >DroYak_CAF1 102109 120 + 1 UACGCGAUUGGUUAGCGUUAAGUGGACGCCAAUCGCAUUUGCUGAUCGGAGCAGCGUAAUUUCCGCCUAAUUAUGUUGACCUAAUGAUUUUGAAAACUACUCAUGCUGAUUUUUAAAAAG ...(((((((((..((.....))....))))))))).((((..((((((..((((((((((.......)))))))))).....((((.............)))).))))))..))))... ( -30.72) >consensus UACGCGAUUGGUUAGCGUUAAGUGGACGCCAAUCGCAUUUGGUGAUCAGAGCAGAGUAAUUCCCGCCUAAUUAUAUUAACUUAAUGGUUUUAAAAGCAACUCAAGCCGAUUAUUAAAAAU ...(((((((((..((.....))....))))))))).((((((((((((.((............)))).................(((((............)))))))))))))))... (-22.90 = -23.06 + 0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:23 2006