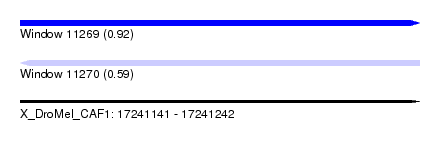
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,241,141 – 17,241,242 |
| Length | 101 |
| Max. P | 0.919510 |

| Location | 17,241,141 – 17,241,242 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -16.02 |
| Energy contribution | -17.22 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17241141 101 + 22224390 -----------------GCGGGGGAUCCG-GGGAAUCGCCUGGCAAAUUGGCCGGCGCAGAAAUGUGUCGGAAGUCUUUAGUAAAAGUUAAAUAGAAAGCAUAUAGGAGAUUUAUAUGC -----------------((..((((((((-((......)))))........((((((((....))))))))..)))))..))................((((((((.....)))))))) ( -31.00) >DroVir_CAF1 104782 91 + 1 --------AGGGG-UA------G---CCG------AC----GGCAUCGGCGUCGGCUCAGAAAUGUGUCGGAAGUCUUUAGUAAAAGUUAAAUAGAAAGCAUAUAGAAGAUUUAUAUGC --------(((((-..------(---(((------(.----....))))).(((((.((....)).)))))...)))))...................(((((((((...))))))))) ( -23.00) >DroGri_CAF1 110912 103 + 1 --------AGGGGGUAAAUUGAGCA-CCG---UAUUC----GCCAUAGUCGUCGACUCUGAAAUGUGUCGGAAGUCUUUAGUAAAAGUUAAAUAGAAAGCAUAUAAAAGAUUUAUAUGC --------....((..(((((....-.))---.))).----.)).........((((((((......)))).))))......................((((((((.....)))))))) ( -17.00) >DroSim_CAF1 89196 101 + 1 -----------------ACGGGGGAUCCG-GGGAAUCGCCUGGCAAAUUGGCCGGCGCAGAAAUGUGUCGGAAGUCUUUAGUAAAAGUUAAAUAGAAAGCAUAUAGGAGAUUUAUAUGC -----------------.((((.((((..-..).))).))))(((......((((((((....))))))))(((((((..(((...(((........))).)))..)))))))...))) ( -31.30) >DroEre_CAF1 93253 99 + 1 G--------------------GGAAUCCGGGGGAAUCGCCUGGCAAAUUGGCCGGCGCAGAAAUGUGUCGGAAGUCUUUAGUAAAAGCUAAAUAGAAAGCAUAUGGGAGAUUUAUAUGC .--------------------.....(((((.(...).)))))........((((((((....))))))))(((((((..(((...(((........))).)))..)))))))...... ( -29.40) >DroYak_CAF1 95985 117 + 1 GGGGUAUACGGGG-UAUACGGGGAAUACG-GGGAAUCGCCUGGCAAAUUGGCCGGCGCAGAAAUGUGUCGGAAGUGUUUAGUAAAAGUUAAAUAGAAAGCAUAUGGAAGAUUUAUAUGC ...((((((...)-)))))...(((((((-((......)))..........((((((((....))))))))..))))))...................(((((((((...))))))))) ( -31.00) >consensus _________________ACGGGGAAUCCG_GGGAAUCGCCUGGCAAAUUGGCCGGCGCAGAAAUGUGUCGGAAGUCUUUAGUAAAAGUUAAAUAGAAAGCAUAUAGAAGAUUUAUAUGC ...................................................((((((((....))))))))...........................(((((((((...))))))))) (-16.02 = -17.22 + 1.20)

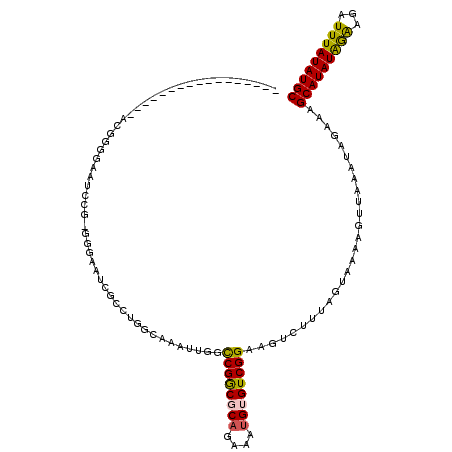
| Location | 17,241,141 – 17,241,242 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -19.24 |
| Consensus MFE | -8.49 |
| Energy contribution | -10.99 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17241141 101 - 22224390 GCAUAUAAAUCUCCUAUAUGCUUUCUAUUUAACUUUUACUAAAGACUUCCGACACAUUUCUGCGCCGGCCAAUUUGCCAGGCGAUUCCC-CGGAUCCCCCGC----------------- (((((((.......)))))))...........((((....))))...((((.((......))(((((((......))).))))......-))))........----------------- ( -20.90) >DroVir_CAF1 104782 91 - 1 GCAUAUAAAUCUUCUAUAUGCUUUCUAUUUAACUUUUACUAAAGACUUCCGACACAUUUCUGAGCCGACGCCGAUGCC----GU------CGG---C------UA-CCCCU-------- (((((((.......)))))))...........((((....))))..................((((((((.......)----))------)))---)------).-.....-------- ( -20.40) >DroGri_CAF1 110912 103 - 1 GCAUAUAAAUCUUUUAUAUGCUUUCUAUUUAACUUUUACUAAAGACUUCCGACACAUUUCAGAGUCGACGACUAUGGC----GAAUA---CGG-UGCUCAAUUUACCCCCU-------- ((((((((.....))))))))...........((((....))))......(((((...(((.((((...)))).)))(----(....---)))-)).))............-------- ( -14.50) >DroSim_CAF1 89196 101 - 1 GCAUAUAAAUCUCCUAUAUGCUUUCUAUUUAACUUUUACUAAAGACUUCCGACACAUUUCUGCGCCGGCCAAUUUGCCAGGCGAUUCCC-CGGAUCCCCCGU----------------- (((((((.......)))))))...........((((....))))...((((.((......))(((((((......))).))))......-))))........----------------- ( -20.90) >DroEre_CAF1 93253 99 - 1 GCAUAUAAAUCUCCCAUAUGCUUUCUAUUUAGCUUUUACUAAAGACUUCCGACACAUUUCUGCGCCGGCCAAUUUGCCAGGCGAUUCCCCCGGAUUCC--------------------C ((((((.........))))))......(((((......)))))((..((((...........(((((((......))).)))).......)))).)).--------------------. ( -20.07) >DroYak_CAF1 95985 117 - 1 GCAUAUAAAUCUUCCAUAUGCUUUCUAUUUAACUUUUACUAAACACUUCCGACACAUUUCUGCGCCGGCCAAUUUGCCAGGCGAUUCCC-CGUAUUCCCCGUAUA-CCCCGUAUACCCC ((((((.........)))))).........................................(((((((......))).))))......-..........(((((-(...))))))... ( -18.70) >consensus GCAUAUAAAUCUCCUAUAUGCUUUCUAUUUAACUUUUACUAAAGACUUCCGACACAUUUCUGCGCCGGCCAAUUUGCCAGGCGAUUCCC_CGGAUCCCCCGU_________________ (((((((.......)))))))..........................((((...........(((((((......))).)))).......))))......................... ( -8.49 = -10.99 + 2.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:16 2006