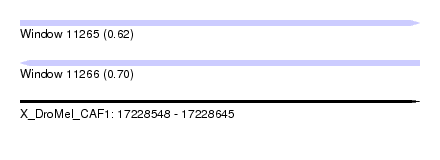
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,228,548 – 17,228,645 |
| Length | 97 |
| Max. P | 0.699968 |

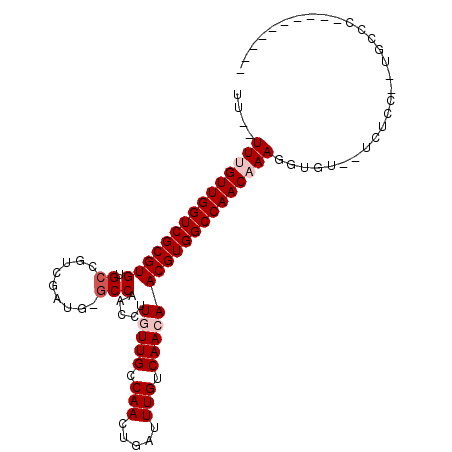
| Location | 17,228,548 – 17,228,645 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -20.68 |
| Energy contribution | -20.82 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17228548 97 + 22224390 GGGGUUACG-GGGCA--GGAGA--ACACCUUUGUUGGCCACGUUGUUGACAAAUCAUUUGGCAAGAACGUUGGC-CAUCGACGGCACACGCGACCAACAAA--AA ..(((..((-..(((--((...--...)))(((.((((((((((.(((.(((.....))).))).)))).))))-)).)))..))...))..)))......--.. ( -32.60) >DroPse_CAF1 103751 90 + 1 ----------GAGAAGGAGAGA--ACACCUUUGUUGGCCACGUUGUUGACAAAUCAGUUGGCAACAAGGUUGGC-CAUCGACGGCACACGCGACCAACGAA--CA ----------..(((((.....--...)))))(((((.(..(((((..((......))..)))))...((((((-(......))).)).))).)))))...--.. ( -27.10) >DroSim_CAF1 76748 91 + 1 ------ACG-GGGCA--GGAGA--ACACCUUUGUUGGCCACGUUGUUGACAAAUCAGUUGGCAAGAACGUUGGC-CAUCGACGGCACACGCGACCAACAAA--AA ------..(-(.((.--(....--.).((.(((.((((((((((.(((.(((.....))).))).)))).))))-)).))).)).....))..))......--.. ( -27.60) >DroYak_CAF1 82751 100 + 1 CGGGUUAAGGGGGCA--GGAGA--ACACCUUUGUUGGCCACGUUGUUGACAAAUCAGUUGGCAACAACGUUGGC-CAUCGACGGCACACGCGACCAACAAAAAAA ..((((..(...(((--((...--...)))(((.((((((((((((((.(((.....))).)))))))).))))-)).)))..))...)..)))).......... ( -33.80) >DroAna_CAF1 107045 80 + 1 --------------A--AGAGAAGAGACCUUUGUUGGCCACGUUGUUGACAAAUCAGUUGGCAACAAGGCUGG-------ACGGCACACGCGACCAACAAA--AA --------------.--............((((((((.(.((((((..((......))..)))))...(((..-------..)))....).).))))))))--.. ( -23.90) >DroPer_CAF1 97492 91 + 1 ----------GAGAAGGAGAGA--ACACCUUUGUUGGCCACGUUGUUGACAAAUCAGUUGGCAACAAGGUUGGCCCAUCGACGGCACACGCGACCAACGAA--CA ----------..(((((.....--...)))))((..(((..(((((..((......))..)))))..)))..))...(((.((.....)))))........--.. ( -26.80) >consensus __________GGGCA__AGAGA__ACACCUUUGUUGGCCACGUUGUUGACAAAUCAGUUGGCAACAACGUUGGC_CAUCGACGGCACACGCGACCAACAAA__AA .............................((((((((.(.((((((..((......))..)))))...((((.........))))....).).)))))))).... (-20.68 = -20.82 + 0.14)

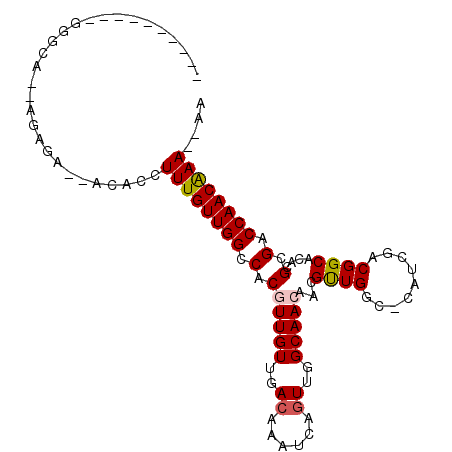
| Location | 17,228,548 – 17,228,645 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -21.60 |
| Energy contribution | -22.43 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17228548 97 - 22224390 UU--UUUGUUGGUCGCGUGUGCCGUCGAUG-GCCAACGUUCUUGCCAAAUGAUUUGUCAACAACGUGGCCAACAAAGGUGU--UCUCC--UGCCC-CGUAACCCC ..--((((((((((((((..((((....))-))..))(((.(((.(((.....))).))).)))))))))))))))((.((--.....--.)).)-)........ ( -29.50) >DroPse_CAF1 103751 90 - 1 UG--UUCGUUGGUCGCGUGUGCCGUCGAUG-GCCAACCUUGUUGCCAACUGAUUUGUCAACAACGUGGCCAACAAAGGUGU--UCUCUCCUUCUC---------- ..--...((((((((((((.((((....))-))).....(((((.(((.....))).)))))))))))))))).((((.(.--...).))))...---------- ( -30.30) >DroSim_CAF1 76748 91 - 1 UU--UUUGUUGGUCGCGUGUGCCGUCGAUG-GCCAACGUUCUUGCCAACUGAUUUGUCAACAACGUGGCCAACAAAGGUGU--UCUCC--UGCCC-CGU------ ..--((((((((((((((.(((.((((.((-((..........))))..))))..).))...))))))))))))))((.((--.....--.)).)-)..------ ( -29.40) >DroYak_CAF1 82751 100 - 1 UUUUUUUGUUGGUCGCGUGUGCCGUCGAUG-GCCAACGUUGUUGCCAACUGAUUUGUCAACAACGUGGCCAACAAAGGUGU--UCUCC--UGCCCCCUUAACCCG ....((((((((((((((..((((....))-))..))(((((((.(((.....))).)))))))))))))))))))(((..--.....--.)))........... ( -34.70) >DroAna_CAF1 107045 80 - 1 UU--UUUGUUGGUCGCGUGUGCCGU-------CCAGCCUUGUUGCCAACUGAUUUGUCAACAACGUGGCCAACAAAGGUCUCUUCUCU--U-------------- ((--((((((((((((((..((...-------...))..(((((.(((.....))).)))))))))))))))))))))..........--.-------------- ( -26.80) >DroPer_CAF1 97492 91 - 1 UG--UUCGUUGGUCGCGUGUGCCGUCGAUGGGCCAACCUUGUUGCCAACUGAUUUGUCAACAACGUGGCCAACAAAGGUGU--UCUCUCCUUCUC---------- ..--...((((((((((((.(((.......)))).....(((((.(((.....))).)))))))))))))))).((((.(.--...).))))...---------- ( -29.90) >consensus UU__UUUGUUGGUCGCGUGUGCCGUCGAUG_GCCAACCUUGUUGCCAACUGAUUUGUCAACAACGUGGCCAACAAAGGUGU__UCUCC__UGCCC__________ ....(((((((((((((((.((.........))).....(((((.(((.....))).)))))))))))))))))))............................. (-21.60 = -22.43 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:12 2006