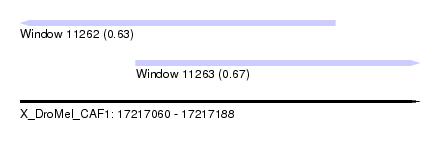
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,217,060 – 17,217,188 |
| Length | 128 |
| Max. P | 0.665392 |

| Location | 17,217,060 – 17,217,161 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.07 |
| Mean single sequence MFE | -18.04 |
| Consensus MFE | -15.40 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
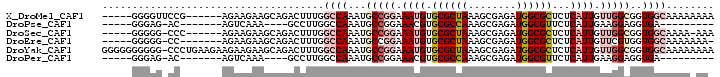
>X_DroMel_CAF1 17217060 101 - 22224390 GCGCCAUCUCGCUUUAGCGCACAUUUCCGGCAUUUGGCCAAAGUCUGCUUCUUCU------CGGAACCCC-------CCAUCC-CCCCCCCCCCCCUUCCACUUUCUUGGCUAUC ..((((....((....))(((.((((..(((.....))).)))).))).......------.(((.....-------...)))-.......................)))).... ( -16.80) >DroSec_CAF1 62872 90 - 1 GCGCCAUCUCGCUUUAGCGCACAUUUCCGGCAUUUGGCCAAAGUCUGCUUCUUCU------GGG-CCCCC-------ACAUUU-CCCC----------CCACUUUCUUGGCCAUC ..(((....(((....))).........)))...((((((((((..........(------((.-...))-------).....-....----------..)))...))))))).. ( -18.32) >DroEre_CAF1 69917 88 - 1 GCGCCAUCUCGCUUUAGCGCACAUUUCCGGCAUUUGGCCAAAGUCUGCUUCUUCU-------GG-CCCCC-------ACAUUU--CCC----------CCACUUUCUUGGCCAUC ((((.(........).))))........(((....((((((((.......))).)-------))-))...-------......--...----------.((......)))))... ( -18.20) >DroYak_CAF1 70718 104 - 1 GCGCCAUCUCGCUUUAGCGCACAUUUCCGGCAUUUGGCCAAAGUCUGCUUCUUCUUCUUCAGGG-CCCCCCCCCCCAACAUUUUCCCC----------CCACUUUCUUGGCCAUC ..(((....(((....))).........)))...(((((((((....)))...........(((-........)))............----------.........)))))).. ( -18.82) >consensus GCGCCAUCUCGCUUUAGCGCACAUUUCCGGCAUUUGGCCAAAGUCUGCUUCUUCU______GGG_CCCCC_______ACAUUU_CCCC__________CCACUUUCUUGGCCAUC ..((((....((....))(((.((((..(((.....))).)))).)))...........................................................)))).... (-15.40 = -15.40 + 0.00)



| Location | 17,217,097 – 17,217,188 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -16.74 |
| Energy contribution | -18.38 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17217097 91 + 22224390 -----GGGGUUCCG------AGAAGAAGCAGACUUUGGCCAAAUGCCGGAAAUGUGCGCUAAAGCGAGAUGGCGCUCUCAUUGUUGGCGGUGGCAAAAAAAA -----..(.(((..------....))).)........((((..((((((.((((.((((((........))))))...)))).)))))).))))........ ( -27.00) >DroPse_CAF1 89385 76 + 1 -----GGGAG-AC-------AGUCAAA----GCCUUGGCCAAAUGCCGGAAACGUGCGCCAAAGCGAGAUGGCGUUCUCAUUGAAGGAGGUGA--------- -----(((((-..-------.((((..----((.(((((.....((((....)).))))))).))....)))).)))))..............--------- ( -23.70) >DroSec_CAF1 62899 89 + 1 -----GGGGG-CCC------AGAAGAAGCAGACUUUGGCCAAAUGCCGGAAAUGUGCGCUAAAGCGAGAUGGCGCUCUCAUUGUUGGCGGUGGCAAAA-AAA -----((...-.))------....((((....)))).((((..((((((.((((.((((((........))))))...)))).)))))).))))....-... ( -28.00) >DroEre_CAF1 69943 88 + 1 -----GGGGG-CC-------AGAAGAAGCAGACUUUGGCCAAAUGCCGGAAAUGUGCGCUAAAGCGAGAUGGCGCUCUCAUUGUUCGUGGUGGCAAAAAAA- -----...((-((-------(((..........)))))))...(((((((((((.((((((........))))))...)))..)))....)))))......- ( -24.80) >DroYak_CAF1 70748 101 + 1 GGGGGGGGGG-CCCUGAAGAAGAAGAAGCAGACUUUGGCCAAAUGCCGGAAAUGUGCGCUAAAGCGAGAUGGCGCUCUCAUUGUUGGCGGUGGCAAAAAAAA ..((((....-)))).........((((....)))).((((..((((((.((((.((((((........))))))...)))).)))))).))))........ ( -30.80) >DroPer_CAF1 83948 76 + 1 -----GGGAG-AC-------AGUCAAA----GCCUUGGCCAAAUGCCGGAAACGUGCGCCAAAGCGAGAUGGCGUUCUCAUUGAAGGAGGUGA--------- -----(((((-..-------.((((..----((.(((((.....((((....)).))))))).))....)))).)))))..............--------- ( -23.70) >consensus _____GGGGG_CC_______AGAAGAAGCAGACUUUGGCCAAAUGCCGGAAAUGUGCGCUAAAGCGAGAUGGCGCUCUCAUUGUUGGCGGUGGCAAAA_AA_ .....................................((((...(((((.((((.((((((........))))))...)))).)))))..))))........ (-16.74 = -18.38 + 1.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:09 2006