
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,210,600 – 17,210,707 |
| Length | 107 |
| Max. P | 0.578341 |

| Location | 17,210,600 – 17,210,707 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.38 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -15.89 |
| Energy contribution | -17.75 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
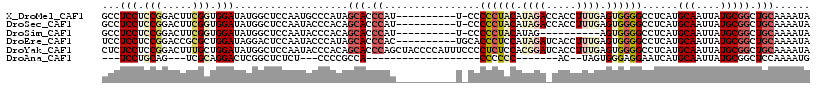
>X_DroMel_CAF1 17210600 107 + 22224390 GCCUCCUCCGGACUUCGGUGGAUAUGGCUCCAAUGCCCAUAGCACCCAU----------U-CCCCCUACAUAGACCACCUUUGAGUGGGGCCUCAUGCAAUUAUGCGGCUGCAAAAUA ...(((.(((.....))).)))((((((......).)))))(((.((..----------.-((((((.((.((.....)).)))).))))......(((....))))).)))...... ( -30.20) >DroSec_CAF1 56179 107 + 1 GCCUCCUCCGGACUUCGGUGGAUAUGGCUCCAAUACCCACAGCACCCAU----------U-CCCCCUACAUAGACCACCUUUGAGUGGGGCCUCAUGCAAUUAUGCGGCUGCAAAAUA ((((((.(((.....))).)))...))).............(((.((..----------.-((((((.((.((.....)).)))).))))......(((....))))).)))...... ( -29.60) >DroSim_CAF1 61209 97 + 1 GCCUCCUCCGGACUUCGGUGGAUAUGGCUCCAAUACCCACAGCACCCAU----------U-CCCCCUACAUAG----------AGUGGGGCCUCAUGCAAUUAUGCGGCUGCAAAAUA ((((((.(((.....))).)))...))).............((((((((----------(-(..........)----------))))))(((.((((....)))).))))))...... ( -30.50) >DroEre_CAF1 62838 108 + 1 UCCUCCUCCGGACCGCGCUGGAUAGGACUCCAAUACCCAUAGCACCCAC----------UGCACCCUCCAUAGAUCACCUUUGAGUGGGGCCUCAUGCAAUUAUGCGGCUGCAAAAUA ((((..(((((......))))).))))..............((((((((----------(.((..................))))))))(((.((((....)))).))))))...... ( -27.37) >DroYak_CAF1 63308 118 + 1 CUCUCCUCCGGACUUUGCUGGAUAUGGCUCCAAUACCCACAGCACCCAGCUACCCCAUUUCCCCUCUCCACGGAUCACCUUUGAGUGGGGCCUCAUGCAAUUAUGCGGCUGCAAAAUA .........((.....(((((.....(((...........)))..)))))....)).....(((.(((...((....))...))).)))(((.((((....)))).)))......... ( -26.90) >DroAna_CAF1 85950 81 + 1 ---UCCUGCAG---UCGCAGGACUCGGCUCUCU---CCCCGCCA-------------------CCCCCC-------AC--UAGUGGGAGGAAUCAUGCAAUUAUGCGGCUCCAAAAUG ---((((((..---..))))))...((......---..))(((.-------------------((.(((-------(.--...)))).))......(((....))))))......... ( -26.30) >consensus GCCUCCUCCGGACUUCGCUGGAUAUGGCUCCAAUACCCACAGCACCCAU__________U_CCCCCUACAUAGA_CACCUUUGAGUGGGGCCUCAUGCAAUUAUGCGGCUGCAAAAUA ...(((.(((.....))).)))...................(((.((................((((((.((((.....)))).))))))......(((....))))).)))...... (-15.89 = -17.75 + 1.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:07 2006