| Sequence ID | X_DroMel_CAF1 |
|---|---|
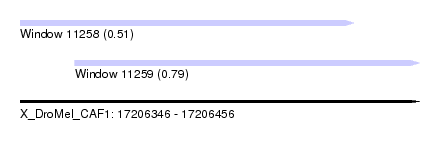
| Location | 17,206,346 – 17,206,456 |
| Length | 110 |
| Max. P | 0.789171 |

| Location | 17,206,346 – 17,206,438 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17206346 92 + 22224390 CCGAUAUCAGUGUA--------UAUACACGUAGAUCUACUUGGGCCCUGCUAAUUAUUCAUCGGGUUCAUCACAUCGUGUGAACUCGGUUUGUAGGUUUU ((((.....((((.--------...))))(((....)))))))..(((((.........((((((((((.(((...)))))))))))))..))))).... ( -24.50) >DroSec_CAF1 51922 92 + 1 CCGAUAUCAGUGUA--------UAUACACGUAGAUCUACUUGGGCCCUGCUAAUUAUUCAUCGGGUUCAUCACAUCGUGUGAACUCGGUUUGUAGGUUUU ((((.....((((.--------...))))(((....)))))))..(((((.........((((((((((.(((...)))))))))))))..))))).... ( -24.50) >DroSim_CAF1 56923 92 + 1 CCGAUAUCAGUGUA--------UAUACACGUAGAUCUACUUGGGCCCUGCUAAUUAUUCAUCGGGUUCAUCACAUCGUGUGAACUCGGUUUGUAGGUUUU ((((.....((((.--------...))))(((....)))))))..(((((.........((((((((((.(((...)))))))))))))..))))).... ( -24.50) >DroEre_CAF1 58816 92 + 1 CCGAUAUCAGAGUA--------UAUACACGUAGAUCUACUUGGGCCCUGCUAAUUAUUCAUCGGGUUCAUCACAUCGUGUGAACUCGGUUUGUAGGUUUU ((((....(((...--------(((....)))..)))..))))..(((((.........((((((((((.(((...)))))))))))))..))))).... ( -21.00) >DroYak_CAF1 58717 100 + 1 CCGAUAUCAGUGUAUAAAAUCAUAUACACGUACAUCUGCUUGGGCCCUGCUAAUUAUUCAUCGGGUUCAUCACAUCGUGUGAACUCGGUUUGUAGGUUUU ((((.....(((((((......)))))))(((....)))))))..(((((.........((((((((((.(((...)))))))))))))..))))).... ( -27.20) >DroAna_CAF1 80801 84 + 1 CAGAUAUCAGGG--------------CACUUGAAUCUGCUGUGG--CUGCUAAUUAUUCAUCGGGUUCAUCACGUCGUGUGAACUCGGUUUGUAGGUUUU .((...(((.((--------------((........)))).)))--...))..((((..((((((((((.(((...)))))))))))))..))))..... ( -22.70) >consensus CCGAUAUCAGUGUA________UAUACACGUAGAUCUACUUGGGCCCUGCUAAUUAUUCAUCGGGUUCAUCACAUCGUGUGAACUCGGUUUGUAGGUUUU .........(((((..........)))))..((((((((...(((...)))........((((((((((.(((...)))))))))))))..)))))))). (-20.52 = -20.80 + 0.28)



| Location | 17,206,361 – 17,206,456 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.20 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -16.72 |
| Energy contribution | -16.88 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17206361 95 + 22224390 AUACACGUAGAUCUACUUGGGCCCUGCUAAUUAUUCAUCGGGUUCAUCACAUCGUGUGAACUCGGUUUGUAGGUUUUUGUUAUAAGUUGUUCGUU---------------------- ....(((.((((((((...(((...)))........((((((((((.(((...)))))))))))))..)))))))).)))...............---------------------- ( -22.60) >DroPse_CAF1 76523 85 + 1 --------ACAUC----UAGGGCCUGCUAAUUAUUCAUCGGGUUCAUCACAUCGUGUGAACUCGGUUUGUAGGUUUUUGUUAUAAGA-GUAUGCUGUU------------------- --------.....----.(((((((((.........((((((((((.(((...)))))))))))))..)))))))))..........-..........------------------- ( -20.70) >DroEre_CAF1 58831 95 + 1 AUACACGUAGAUCUACUUGGGCCCUGCUAAUUAUUCAUCGGGUUCAUCACAUCGUGUGAACUCGGUUUGUAGGUUUUUGUUAUAAGUUGUUCGUU---------------------- ....(((.((((((((...(((...)))........((((((((((.(((...)))))))))))))..)))))))).)))...............---------------------- ( -22.60) >DroYak_CAF1 58740 95 + 1 AUACACGUACAUCUGCUUGGGCCCUGCUAAUUAUUCAUCGGGUUCAUCACAUCGUGUGAACUCGGUUUGUAGGUUUUUGUUAUAAGUUGUUCGUU---------------------- ....(((.(((.((((..(((((.(((.........((((((((((.(((...)))))))))))))..))))))))..))....)).))).))).---------------------- ( -22.90) >DroAna_CAF1 80813 112 + 1 ---CACUUGAAUCUGCUGUGG--CUGCUAAUUAUUCAUCGGGUUCAUCACGUCGUGUGAACUCGGUUUGUAGGUUUUUGUUAUAAGCAGUUUUUUACUUACUACUUUUUUGCACGUU ---....((((.(((((((((--(......((((..((((((((((.(((...)))))))))))))..))))......))))).)))))...))))..................... ( -27.00) >DroPer_CAF1 71357 85 + 1 --------ACAUC----UGGGGCCUGCUAAUUAUUCAUCGGGUUCAUCACAUCGUGUGAACUCGGUUUGUAGGUUUUUGUUAUAAGA-GUAUGCUGUU------------------- --------....(----..((((((((.........((((((((((.(((...)))))))))))))..))))))))..)........-..........------------------- ( -20.80) >consensus ___CACGUACAUCU_CUUGGGCCCUGCUAAUUAUUCAUCGGGUUCAUCACAUCGUGUGAACUCGGUUUGUAGGUUUUUGUUAUAAGUUGUUCGUU______________________ ......................(((((.........((((((((((.(((...)))))))))))))..)))))............................................ (-16.72 = -16.88 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:06 2006