
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,156,689 – 17,156,806 |
| Length | 117 |
| Max. P | 0.895473 |

| Location | 17,156,689 – 17,156,806 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -49.25 |
| Consensus MFE | -30.55 |
| Energy contribution | -29.92 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17156689 117 + 22224390 CCUAUUUGAGCGCCUGGCCAUACGCGGCCGCCGCUGCCGCCCAAUCGCCCCAC---GGGGCCACGCCCUCCGCGAUCGAUAUCUAUUACCGCCAGGCAGCCGCUGCAGCGGCCAUGCAAA .........(((((((((.....(((((.......)))))...(((((((...---.))))..(((.....)))...)))..........))))))).(((((....)))))...))... ( -49.10) >DroVir_CAF1 2510 117 + 1 CCUAUUUGAGCGCCUGGCCGUAUGCAGCCGCUGCGGCAGCACAGACACCGCAC---GGUGCACCGCCCUCGGCCAUUGAUAUAUACUAUCGCCAAGCGGCAGCAGCCGCCGCACUCCAGA .......(((.((..(((.((.(((.((((((..(((......(.((((....---)))))...(((...)))....((((.....))))))).)))))).))))).))))).))).... ( -42.00) >DroPse_CAF1 14151 117 + 1 CCUAUCUGAGUGCCUGGCCAUAUGCGGCGGCCGCUGCCGCCCAGUCCCCGCAC---GGCGCCCCACCCUCGGCCAUCGAUAUCUACUACCGCCAGGCGGCGGCCGCCGCCGCCCUGCAGA ..........(((..(((.....(((((((((((((((((.........))..---((((((........)))....(....).......))).))))))))))))))).)))..))).. ( -52.60) >DroGri_CAF1 2126 117 + 1 CCUACUUGAGCGCCUGGCCGUAUGCGGCAGCCGCCGCUGCCCAAUCGCCACAC---GGUGCACCUCCCUCGGCGAUUGAUAUUUAUUAUCGGCAAGCGGCCGCAGCCGCCGCGUUGCAGA .....((((((((..((((((.(((((((((....))))))((((((((....---((.......))...)))))))).............))).))))))((....)).))))).))). ( -48.90) >DroWil_CAF1 35328 120 + 1 CCUAUUUGAGUGCUUGGCCAUAUGCAGCCGCUGCUGCGACACAAUCACCGCAUGGCGGUGCCCCGCCCUCAGCAAUUGAUAUCUAUUAUCGUCAGGCAGCAGCGGCAGCUGCCCUGCAAA ..........(((..(((.....((.((((((((((((((.........((..(((((....)))))....))....((((.....)))))))..))))))))))).)).)))..))).. ( -50.30) >DroPer_CAF1 9078 117 + 1 CCUAUCUGAGUGCCUGGCCAUAUGCGGCGGCCGCUGCCGCCCAGUCCCCGCAC---GGCGCCCCACCCUCGGCCAUCGAUAUCUACUACCGCCAGGCGGCGGCCGCCGCCGCCCUGCAGA ..........(((..(((.....(((((((((((((((((.........))..---((((((........)))....(....).......))).))))))))))))))).)))..))).. ( -52.60) >consensus CCUAUUUGAGCGCCUGGCCAUAUGCGGCCGCCGCUGCCGCCCAAUCACCGCAC___GGUGCCCCGCCCUCGGCCAUCGAUAUCUACUACCGCCAGGCGGCAGCAGCCGCCGCCCUGCAGA ....((((..((((((((.....(((((....)))))...........(((......)))..............................))))))))(((((....)))))....)))) (-30.55 = -29.92 + -0.63)



| Location | 17,156,689 – 17,156,806 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -60.03 |
| Consensus MFE | -36.77 |
| Energy contribution | -36.92 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17156689 117 - 22224390 UUUGCAUGGCCGCUGCAGCGGCUGCCUGGCGGUAAUAGAUAUCGAUCGCGGAGGGCGUGGCCCC---GUGGGGCGAUUGGGCGGCAGCGGCGGCCGCGUAUGGCCAGGCGCUCAAAUAGG ...((..((((((....))))))(((((((.(((.......((((((((.....))...((((.---...))))))))))(((((.......))))).))).)))))))))......... ( -57.80) >DroVir_CAF1 2510 117 - 1 UCUGGAGUGCGGCGGCUGCUGCCGCUUGGCGAUAGUAUAUAUCAAUGGCCGAGGGCGGUGCACC---GUGCGGUGUCUGUGCUGCCGCAGCGGCUGCAUACGGCCAGGCGCUCAAAUAGG ....((((((.(((((((((((..((((((((((.....))))....))))))(((((((((((---....)))))....)))))))))))))))))...(.....)))))))....... ( -58.00) >DroPse_CAF1 14151 117 - 1 UCUGCAGGGCGGCGGCGGCCGCCGCCUGGCGGUAGUAGAUAUCGAUGGCCGAGGGUGGGGCGCC---GUGCGGGGACUGGGCGGCAGCGGCCGCCGCAUAUGGCCAGGCACUCAGAUAGG ..(((..(((.(((((((((.((((((..((((....(....)....)))).)))))).(((((---((.((....).).))))).))))))))))).....)))..))).......... ( -63.90) >DroGri_CAF1 2126 117 - 1 UCUGCAACGCGGCGGCUGCGGCCGCUUGCCGAUAAUAAAUAUCAAUCGCCGAGGGAGGUGCACC---GUGUGGCGAUUGGGCAGCGGCGGCUGCCGCAUACGGCCAGGCGCUCAAGUAGG .((((...(((((((((...((((((.(((((((.....))))((((((((.((........))---...)))))))).))))))))))))))))))....(((.....)))...)))). ( -57.30) >DroWil_CAF1 35328 120 - 1 UUUGCAGGGCAGCUGCCGCUGCUGCCUGACGAUAAUAGAUAUCAAUUGCUGAGGGCGGGGCACCGCCAUGCGGUGAUUGUGUCGCAGCAGCGGCUGCAUAUGGCCAAGCACUCAAAUAGG ..(((..(((.((.(((((((((((..(((((((.....))))(((..(((..(((((....)))))...)))..)))..)))))))))))))).)).....)))..))).......... ( -59.30) >DroPer_CAF1 9078 117 - 1 UCUGCAGGGCGGCGGCGGCCGCCGCCUGGCGGUAGUAGAUAUCGAUGGCCGAGGGUGGGGCGCC---GUGCGGGGACUGGGCGGCAGCGGCCGCCGCAUAUGGCCAGGCACUCAGAUAGG ..(((..(((.(((((((((.((((((..((((....(....)....)))).)))))).(((((---((.((....).).))))).))))))))))).....)))..))).......... ( -63.90) >consensus UCUGCAGGGCGGCGGCGGCCGCCGCCUGGCGAUAAUAGAUAUCAAUGGCCGAGGGCGGGGCACC___GUGCGGCGACUGGGCGGCAGCGGCGGCCGCAUAUGGCCAGGCACUCAAAUAGG ((((...((((((....))))))(((((((((((.....))))....((..........((((....))))........((((((....)))))))).....)))))))...)))).... (-36.77 = -36.92 + 0.14)
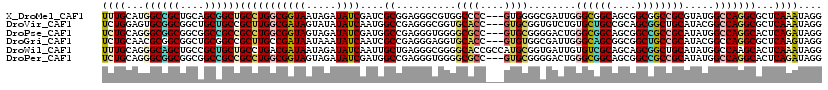

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:51 2006