| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,147,137 – 17,147,250 |
| Length | 113 |
| Max. P | 0.840575 |

| Location | 17,147,137 – 17,147,250 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -27.45 |
| Energy contribution | -28.82 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
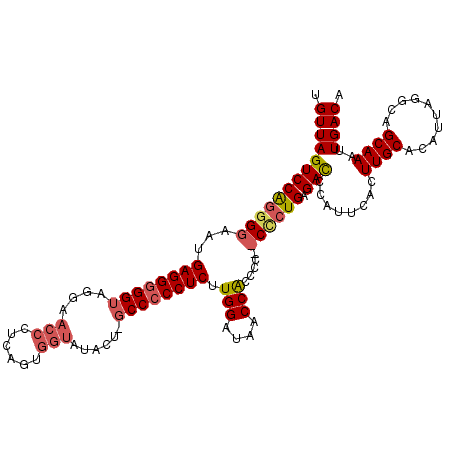
>X_DroMel_CAF1 17147137 113 + 22224390 UGUUAGUCCACAGGAAUGAGGUGGUAGGAACCCCCAGUGGUAUACA-GCCCCCUCUUGGAUAGCCACCCCCCCCCUGAGACCCAUUCACUUGCACAUUAGGCAGCAAAUUGACA .(((((((..((((...(.((((((.((.....))...(((.....-))).((....))...)))))).)...)))).)))........((((..........))))..)))). ( -25.20) >DroSec_CAF1 10195 112 + 1 UGUUAGUCCAGGGGAAUGAGGGGGUAGGAACCCUCAGUGGUAUACU-GCCCCCUCUUGGAAAACCACCCC-CCCCUGAGACCCAUUCACUUGCACAUUAGGCAGCAAAUUGACA .(((((((((((((...((((((((((..(((......)))...))-)))))))).(((....)))....-)))))).)))........((((..........))))..)))). ( -44.50) >DroSim_CAF1 11296 111 + 1 UGUUAGUCCAGGGGAAUGAGGGGGUAUGAACCCUCAGUGGUAUACU-GCCCCCUCUUGGAUAACCACCCC--CCCUGAGACCCAUUCACUUGCACAUUAGGCAGCAAAUUGACA .(((((((((((((...(((((((((((.(((......))).)).)-)))))))).(((....)))...)--))))).)))........((((..........))))..)))). ( -40.90) >DroEre_CAF1 10898 101 + 1 UGUUAGUCCGGGG-GUUGAGGGGGUAGGA---------GGUAGGAGGCCCCCCUCGUGGAUACCCGCCC---CUCUGAGAUCCAUUCACUUGCACAUUAGGCAGCAAAUUGACA .((((((....((-(((.((((((...((---------((..((...))..))))((((....))))))---))))..)))))......((((..........)))))))))). ( -32.10) >consensus UGUUAGUCCAGGGGAAUGAGGGGGUAGGAACCCUCAGUGGUAUACU_GCCCCCUCUUGGAUAACCACCCC__CCCUGAGACCCAUUCACUUGCACAUUAGGCAGCAAAUUGACA .(((((((((((((...((((((((....(((......)))......)))))))).(((....))).....)))))).)))........((((..........))))..)))). (-27.45 = -28.82 + 1.37)


| Location | 17,147,137 – 17,147,250 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -40.96 |
| Consensus MFE | -29.09 |
| Energy contribution | -31.28 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
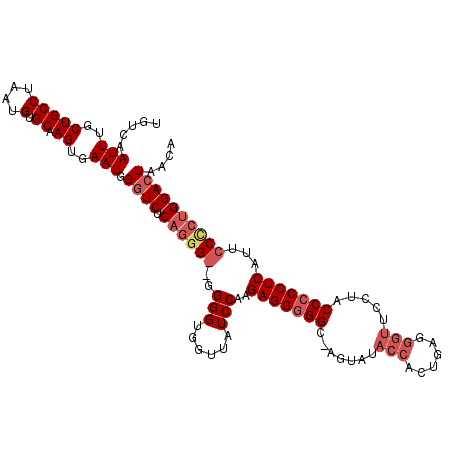
>X_DroMel_CAF1 17147137 113 - 22224390 UGUCAAUUUGCUGCCUAAUGUGCAAGUGAAUGGGUCUCAGGGGGGGGGUGGCUAUCCAAGAGGGGGC-UGUAUACCACUGGGGGUUCCUACCACCUCAUUCCUGUGGACUAACA .....(((..(((((....).)).))..))).(((((((((.(((((((((..((((....((((..-......)).))..))))..))))).))))...)))).))))).... ( -39.50) >DroSec_CAF1 10195 112 - 1 UGUCAAUUUGCUGCCUAAUGUGCAAGUGAAUGGGUCUCAGGGG-GGGGUGGUUUUCCAAGAGGGGGC-AGUAUACCACUGAGGGUUCCUACCCCCUCAUUCCCCUGGACUAACA .....(((..(((((....).)).))..))).((((.((((((-((..(((....))).(((((((.-((...(((......)))..)).))))))).)))))))))))).... ( -46.60) >DroSim_CAF1 11296 111 - 1 UGUCAAUUUGCUGCCUAAUGUGCAAGUGAAUGGGUCUCAGGG--GGGGUGGUUAUCCAAGAGGGGGC-AGUAUACCACUGAGGGUUCAUACCCCCUCAUUCCCCUGGACUAACA .....(((..(((((....).)).))..))).((((.(((((--(((.(((....))).(((((((.-.....(((......))).....))))))).)))))))))))).... ( -44.80) >DroEre_CAF1 10898 101 - 1 UGUCAAUUUGCUGCCUAAUGUGCAAGUGAAUGGAUCUCAGAG---GGGCGGGUAUCCACGAGGGGGGCCUCCUACC---------UCCUACCCCCUCAAC-CCCCGGACUAACA .(((.(((..(((((....).)).))..)))..........(---(((.((....))..(((((((..........---------.....)))))))...-)))).)))..... ( -32.96) >consensus UGUCAAUUUGCUGCCUAAUGUGCAAGUGAAUGGGUCUCAGGG__GGGGUGGUUAUCCAAGAGGGGGC_AGUAUACCACUGAGGGUUCCUACCCCCUCAUUCCCCUGGACUAACA .....(((..(((((....).)).))..))).((((.((((((..(((......)))..(((((((.......(((......))).....)))))))...)))))))))).... (-29.09 = -31.28 + 2.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:50 2006