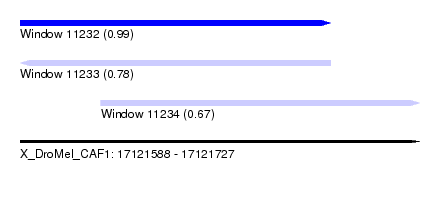
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,121,588 – 17,121,727 |
| Length | 139 |
| Max. P | 0.986737 |

| Location | 17,121,588 – 17,121,696 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.61 |
| Mean single sequence MFE | -20.25 |
| Consensus MFE | -13.71 |
| Energy contribution | -13.03 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17121588 108 + 22224390 AUUAUAUUCUUAUAU-UCUUCUUGUGAUAAAUGGUCUUGCGAGCUAAAAAUUCCCGGUUUUUAUGGCUUGCACUUCUUGUGCAAAGCCGAUUGAUAAACAUAUAUAUUU ..(((((..((((..-((..((((((((.....)))..))))).(((((((.....))))))).(((((((((.....)))).)))))))...))))..)))))..... ( -21.40) >DroSec_CAF1 3976 107 + 1 AUUAUAUUCUUAUAUAUAUUCUUGUGAUAAAUGGUCUUAAGAGUUAAAAAUUC--UAUUUUUAUGGCGUGUACUUCCUGUGCAAAGCUGAUUGAUAAGCAUAUAUAUUU ..(((((.(((((....(((((((.(((.....))).))))))).........--.........(((.(((((.....)))))..))).....))))).)))))..... ( -18.90) >DroEre_CAF1 3798 94 + 1 ----------U---U-CAUUCUUUUGAUAAAUGCUCUUGCGAGUCAAAAAUUCUCAGUUUUGAUAGCUUGCACUUCCUGUGCAGAGCUGAUUGAUAAA-AAAUAUCGUU ----------.---(-((......))).....((((..((..(((((((........))))))).)).(((((.....))))))))).....((((..-...))))... ( -20.40) >DroYak_CAF1 3985 85 + 1 CU--------U---U-CCUUCUUUUGAUAA-----------AGUUAAAAACUCUCAGUUUUUAUGGCUUGCACUUCCUGUGCAGAGCCAAUUGAUAAA-AAAUAUCUUU ..--------.---.-.........((((.-----------...(((((((.....)))))))((((((((((.....))))).))))).........-...))))... ( -20.30) >consensus AU________U___U_CAUUCUUGUGAUAAAUGGUCUUGCGAGUUAAAAAUUCUCAGUUUUUAUGGCUUGCACUUCCUGUGCAAAGCCGAUUGAUAAA_AAAUAUAUUU ............................................(((((((.....)))))))((((((((((.....)))).)))))).................... (-13.71 = -13.03 + -0.69)


| Location | 17,121,588 – 17,121,696 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 72.61 |
| Mean single sequence MFE | -18.38 |
| Consensus MFE | -9.36 |
| Energy contribution | -10.30 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17121588 108 - 22224390 AAAUAUAUAUGUUUAUCAAUCGGCUUUGCACAAGAAGUGCAAGCCAUAAAAACCGGGAAUUUUUAGCUCGCAAGACCAUUUAUCACAAGAAGA-AUAUAAGAAUAUAAU .......(((((((.((....(((((.((((.....)))))))))....................(.((....)).)...........)).))-))))).......... ( -22.00) >DroSec_CAF1 3976 107 - 1 AAAUAUAUAUGCUUAUCAAUCAGCUUUGCACAGGAAGUACACGCCAUAAAAAUA--GAAUUUUUAACUCUUAAGACCAUUUAUCACAAGAAUAUAUAUAAGAAUAUAAU ..((((((((.(((........(((((......)))))........(((((((.--..))))))).....................))).))))))))........... ( -9.20) >DroEre_CAF1 3798 94 - 1 AACGAUAUUU-UUUAUCAAUCAGCUCUGCACAGGAAGUGCAAGCUAUCAAAACUGAGAAUUUUUGACUCGCAAGAGCAUUUAUCAAAAGAAUG-A---A---------- ...((((...-..))))....((((.(((((.....))))))))).((((((........))))))(((....)))(((((.......)))))-.---.---------- ( -21.60) >DroYak_CAF1 3985 85 - 1 AAAGAUAUUU-UUUAUCAAUUGGCUCUGCACAGGAAGUGCAAGCCAUAAAAACUGAGAGUUUUUAACU-----------UUAUCAAAAGAAGG-A---A--------AG ...((((...-..))))...(((((.(((((.....))))))))))(((((((.....))))))).((-----------((.......)))).-.---.--------.. ( -20.70) >consensus AAAGAUAUAU_UUUAUCAAUCAGCUCUGCACAGGAAGUGCAAGCCAUAAAAACUGAGAAUUUUUAACUCGCAAGACCAUUUAUCAAAAGAAGA_A___A________AU ...............((....((((.(((((.....))))))))).(((((((.....))))))).......................))................... ( -9.36 = -10.30 + 0.94)

| Location | 17,121,616 – 17,121,727 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -15.92 |
| Energy contribution | -15.28 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17121616 111 + 22224390 AAUGGUCUUGCGAGCUAAAAAUUCCCGGUUUUUAUGGCUUGCACUUCUUGUGCAAAGCCGAUUGAUAAACAUAUAUAUUUAUAAUU---------GCUUUAAUUAUGCUAUAUAAGUGGC ...((((........(((((((.....))))))).(((((((((.....)))).)))))))))......(((.(((((.(((((((---------(...))))))))..))))).))).. ( -23.60) >DroSec_CAF1 4005 107 + 1 AAUGGUCUUAAGAGUUAAAAAUUC--UAUUUUUAUGGCGUGUACUUCCUGUGCAAAGCUGAUUGAUAAGCAUAUAUAUUUAUAAUU---------GCUUUAAUUAUGCUA--UAAAUGGC ....(((...(((((.....))))--)...((((((((((((((.....)))))....(((((((..((((((((....))))..)---------)))))))))))))))--)))).))) ( -21.20) >DroSim_CAF1 3981 109 + 1 UAUCGUCUAAAGAGUUAAAAAUUCUCUGUUUUUAUGGCUUGCACUUCCUGUGCAAAGCUGAUUGAUAAGCAUAUAUAUUUAUAAUU---------GCUUUAAUUAUGCUA--UAAAUGGC ....(((...((((.........))))...((((((((((((((.....))))))...(((((((..((((((((....))))..)---------)))))))))).))))--)))).))) ( -24.20) >DroEre_CAF1 3813 117 + 1 AAUGCUCUUGCGAGUCAAAAAUUCUCAGUUUUGAUAGCUUGCACUUCCUGUGCAGAGCUGAUUGAUAAA-AAAUAUCGUUAUAAUUCUAAGAAGUGCUUUGCUUAUGCUA--UAAAUGGC ...((....(((((.((....((((.((..(((.((((((((((.....))))).)))))...((((..-...))))....)))..)).)))).)).)))))....))..--........ ( -26.30) >DroYak_CAF1 4002 106 + 1 A-----------AGUUAAAAACUCUCAGUUUUUAUGGCUUGCACUUCCUGUGCAGAGCCAAUUGAUAAA-AAAUAUCUUUACAAUUGCAAGAACUGCUUUACUUAUGCUA--UAAAUGGC .-----------(((..........((((((((.((((((((((.....))))).)))))...((((..-...))))...........))))))))..........))).--........ ( -23.35) >consensus AAUGGUCUUAAGAGUUAAAAAUUCUCAGUUUUUAUGGCUUGCACUUCCUGUGCAAAGCUGAUUGAUAAACAUAUAUAUUUAUAAUU_________GCUUUAAUUAUGCUA__UAAAUGGC ...........(((((((((((.....)))))))((((((((((.....)))).))))))...................................)))).......((((......)))) (-15.92 = -15.28 + -0.64)

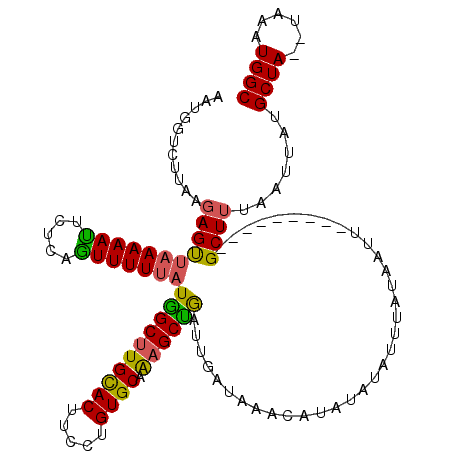
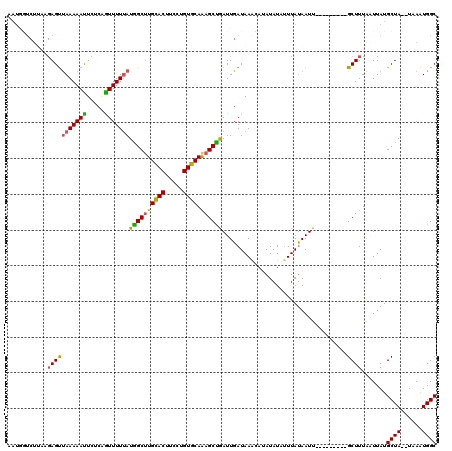
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:43 2006