
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,105,615 – 17,105,725 |
| Length | 110 |
| Max. P | 0.926944 |

| Location | 17,105,615 – 17,105,725 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -47.57 |
| Consensus MFE | -36.04 |
| Energy contribution | -38.10 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17105615 110 + 22224390 GAUCAAGUCCAAGUCCUCCUCGACUUUGAGCUCCGACGUGGAGCCAUUUUACUUGCAUCCACCGGCCUUAGCAGG---CGGAUCAGGAGGAUCGGGAAUGGGCUUGGGCAUGG------- ...((.((((((((((..((((((((((.(((((.....))))))).....((((.((((.((.((....)).))---.))))))))))).)))))...)))))))))).)).------- ( -47.90) >DroSec_CAF1 18 110 + 1 GAUCAAGUCCAAGUCCUCCUCGACUUUGAGCUCCGACGUGGAGCCAUUUUACUUGCAUCCACCGGCCUUAGCAGG---CGGAUCGGGAGGAUCGGGAAUGGGCUUGGCCAUGG------- (..((((((((..((((((((.....((.(((((.....)))))))......(((.((((.((.((....)).))---.))))))))))))..)))..))))))))..)....------- ( -43.50) >DroEre_CAF1 1 104 + 1 -------------UCCUCCUCGACUUUGAGCUCCGACGUGGAGCCAUUCUACUUGCAUCCACCGGCCAUAGCAGG---CGGAGCAGGAGGAUCGGGCAUGGGCUUGGGCAUGGGGAUGGG -------------.((((((((..((..((((((.....)))(((..(((.(((((.(((.((.((....)).))---.)))))))).)))...)))....)))..))..)))))).)). ( -42.80) >DroYak_CAF1 31 112 + 1 GAUCAAGUCCAAGUCCUCCUCGACACUGAGCUCCGACGUGGAGCCAUUCUACUUGCAUCCACCGGCCAUAGCGGGCGGCGGAGCAGGAGGAUCGGGAAUGGGCUUGGGCAUG-------- ......((((((((((..(((((...((.(((((.....)))))))((((.(((((.(((.(((.((.....)).))).)))))))))))))))))...))))))))))...-------- ( -56.10) >consensus GAUCAAGUCCAAGUCCUCCUCGACUUUGAGCUCCGACGUGGAGCCAUUCUACUUGCAUCCACCGGCCAUAGCAGG___CGGAGCAGGAGGAUCGGGAAUGGGCUUGGGCAUGG_______ ......((((((((((..(((((...((.(((((.....)))))))((((.(((((.(((.((.((....)).))....)))))))))))))))))...))))))))))........... (-36.04 = -38.10 + 2.06)

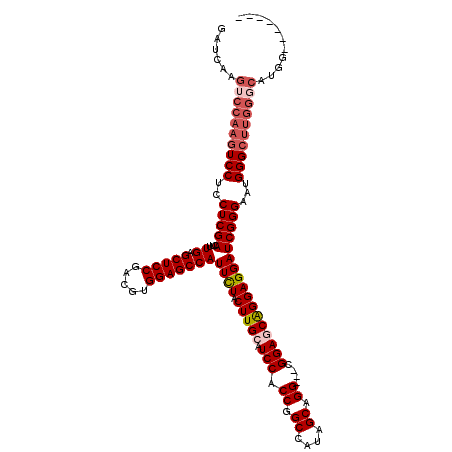

| Location | 17,105,615 – 17,105,725 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -41.83 |
| Consensus MFE | -27.70 |
| Energy contribution | -29.70 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17105615 110 - 22224390 -------CCAUGCCCAAGCCCAUUCCCGAUCCUCCUGAUCCG---CCUGCUAAGGCCGGUGGAUGCAAGUAAAAUGGCUCCACGUCGGAGCUCAAAGUCGAGGAGGACUUGGACUUGAUC -------.......((((.(((.(((...(((((.(((((((---((.((....)).))))))).)).......(((((((.....))))).)).....))))))))..))).))))... ( -41.00) >DroSec_CAF1 18 110 - 1 -------CCAUGGCCAAGCCCAUUCCCGAUCCUCCCGAUCCG---CCUGCUAAGGCCGGUGGAUGCAAGUAAAAUGGCUCCACGUCGGAGCUCAAAGUCGAGGAGGACUUGGACUUGAUC -------....(((...)))((...(((((((((((((((((---((.((....)).))))))...........(((((((.....))))).))...))).)))))).))))...))... ( -41.90) >DroEre_CAF1 1 104 - 1 CCCAUCCCCAUGCCCAAGCCCAUGCCCGAUCCUCCUGCUCCG---CCUGCUAUGGCCGGUGGAUGCAAGUAGAAUGGCUCCACGUCGGAGCUCAAAGUCGAGGAGGA------------- ........((((........)))).....(((((((((((((---((.((....)).)))))).)).....((.(((((((.....))))).))...)).)))))))------------- ( -38.00) >DroYak_CAF1 31 112 - 1 --------CAUGCCCAAGCCCAUUCCCGAUCCUCCUGCUCCGCCGCCCGCUAUGGCCGGUGGAUGCAAGUAGAAUGGCUCCACGUCGGAGCUCAGUGUCGAGGAGGACUUGGACUUGAUC --------......((((.(((.(((...(((((.((((((((((.((.....)).))))))).)))....((.(((((((.....))))).))...))))))))))..))).))))... ( -46.40) >consensus _______CCAUGCCCAAGCCCAUUCCCGAUCCUCCUGAUCCG___CCUGCUAAGGCCGGUGGAUGCAAGUAAAAUGGCUCCACGUCGGAGCUCAAAGUCGAGGAGGACUUGGACUUGAUC .............(((((.((..(((((((......((((((....(.((....)).)(((((.((..........)))))))..)))))).....)))).))))).)))))........ (-27.70 = -29.70 + 2.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:37 2006