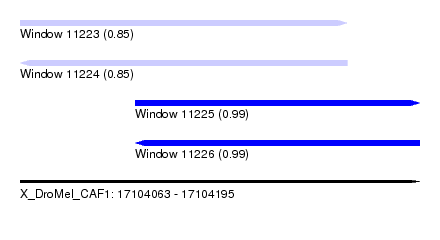
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,104,063 – 17,104,195 |
| Length | 132 |
| Max. P | 0.994163 |

| Location | 17,104,063 – 17,104,171 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 86.90 |
| Mean single sequence MFE | -35.14 |
| Consensus MFE | -24.66 |
| Energy contribution | -24.09 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
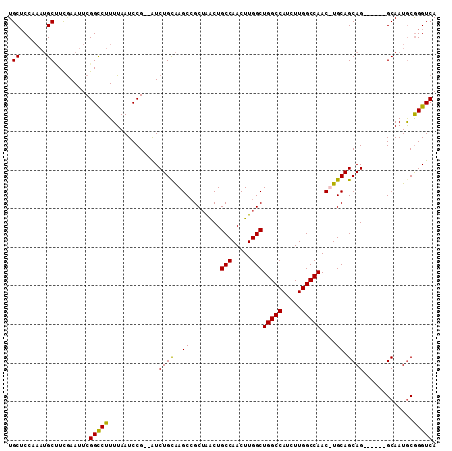
>X_DroMel_CAF1 17104063 108 + 22224390 UGCACCAAAUGCAUCGAAUUCGGUCUUUUAAUCCG--AUCUGUAAGCAGCUAACUGCCAACUUGGCUGGCCAUCUUGGCCAAC-UGCAGCAGUUGAACGCAAUGCAGGUCA .(((.....)))((((....))))..........(--(((((((.((((......(((.....)))(((((.....))))).)-))).((........))..)))))))). ( -34.20) >DroSec_CAF1 110839 103 + 1 UGCUCCAAAUGCCUCGAAUUCGGCCUUUUAAUCCG--AUCUGCAAGCCGCUAACUGCCAACUUGGCUGGCCAUCUUGGCCAACUUGCAGCAG------GCAAUACGGGUCA .((((....(((((.....((((.........)))--).(((((((.........(((.....)))(((((.....))))).))))))).))------)))....)))).. ( -33.90) >DroSim_CAF1 101557 103 + 1 UGCUCCAAAUGCUUCGAAUUCGGCCUUUUAAUCCG--AUCUGCAAGCCGCUAACUGCCAACUUGGCUGGCCAUCUUGGCCAACUUGCAGCAG------GCAAUGCGGGUCA .((.......)).(((....)))...........(--(((((((.((((((....(((.....)))(((((.....)))))......))).)------))..)))))))). ( -33.90) >DroEre_CAF1 101736 100 + 1 UGCUCC----GCUUUAAAUUCGGCCUUUUAAUCCGCAGUCUGCGAGCCGCAGGCUGCCAACUUGGCUGGCCAUCUUGGCCAAC-UGCAGCAG------GCAAUGCGGGUCA .(..((----((.........((((.........((((((((((...))))))))))......))))((((.....))))...-(((.....------)))..))))..). ( -38.56) >consensus UGCUCCAAAUGCUUCGAAUUCGGCCUUUUAAUCCG__AUCUGCAAGCCGCUAACUGCCAACUUGGCUGGCCAUCUUGGCCAAC_UGCAGCAG______GCAAUGCGGGUCA .((.......)).........(((((.............((((..((........(((.....)))(((((.....)))))....)).)))).............))))). (-24.66 = -24.09 + -0.56)

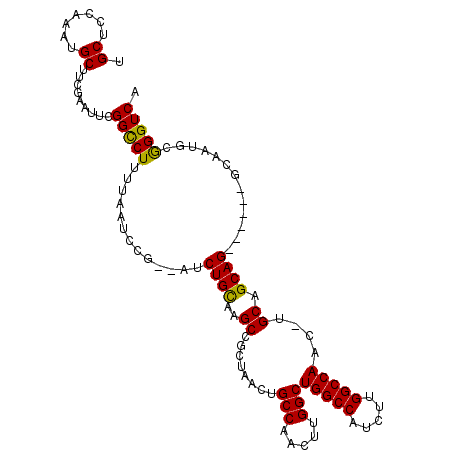
| Location | 17,104,063 – 17,104,171 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 86.90 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -26.73 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17104063 108 - 22224390 UGACCUGCAUUGCGUUCAACUGCUGCA-GUUGGCCAAGAUGGCCAGCCAAGUUGGCAGUUAGCUGCUUACAGAU--CGGAUUAAAAGACCGAAUUCGAUGCAUUUGGUGCA ..((((((((((.((((..(((..(((-(((((((.....)))).(((.....)))....))))))...)))..--.((.........)))))).)))))))...)))... ( -38.20) >DroSec_CAF1 110839 103 - 1 UGACCCGUAUUGC------CUGCUGCAAGUUGGCCAAGAUGGCCAGCCAAGUUGGCAGUUAGCGGCUUGCAGAU--CGGAUUAAAAGGCCGAAUUCGAGGCAUUUGGAGCA .(..(((...(((------((.(((((((((((((.....)))).(((.....))).......))))))))).(--(((.........)))).....)))))..)))..). ( -40.40) >DroSim_CAF1 101557 103 - 1 UGACCCGCAUUGC------CUGCUGCAAGUUGGCCAAGAUGGCCAGCCAAGUUGGCAGUUAGCGGCUUGCAGAU--CGGAUUAAAAGGCCGAAUUCGAAGCAUUUGGAGCA .....((((((((------(.(((....(((((((.....)))))))..))).))))))..)))((((.(((((--((((((.........)))))))....)))))))). ( -36.60) >DroEre_CAF1 101736 100 - 1 UGACCCGCAUUGC------CUGCUGCA-GUUGGCCAAGAUGGCCAGCCAAGUUGGCAGCCUGCGGCUCGCAGACUGCGGAUUAAAAGGCCGAAUUUAAAGC----GGAGCA .(..((((..(((------(.(((...-(((((((.....)))))))..))).))))((((..((..(((.....)))..))...))))..........))----))..). ( -41.20) >consensus UGACCCGCAUUGC______CUGCUGCA_GUUGGCCAAGAUGGCCAGCCAAGUUGGCAGUUAGCGGCUUGCAGAU__CGGAUUAAAAGGCCGAAUUCGAAGCAUUUGGAGCA .(..(((...(((........(((((....(((((.....)))))(((.....))).....))))).....((...(((.........)))...))...)))..)))..). (-26.73 = -27.10 + 0.37)

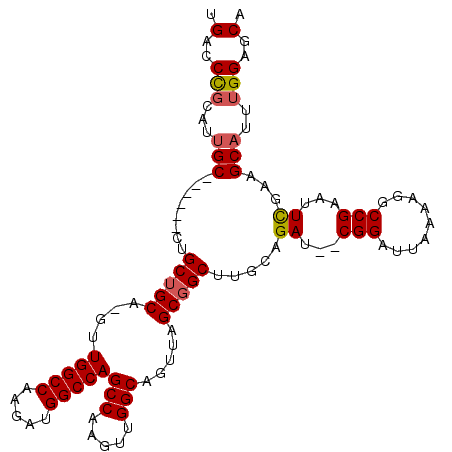
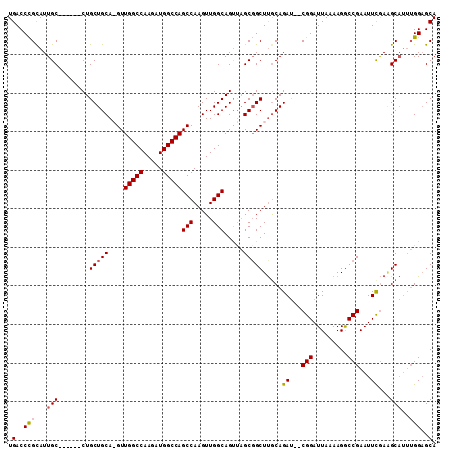
| Location | 17,104,101 – 17,104,195 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -20.66 |
| Energy contribution | -22.10 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17104101 94 + 22224390 UGUAAGCAGCUAACUGCCAACUUGGCUGGCCAUCUUGGCCAAC-UGCAGCAGUUGAACGCAAUGCAGGUCAAUUGCUACGUUAAGUGCAUUGGCC ........(((((.(((..((((((((((((.....)))))..-.(.(((((((((..((...))...))))))))).)))))))))))))))). ( -34.10) >DroSec_CAF1 110877 89 + 1 UGCAAGCCGCUAACUGCCAACUUGGCUGGCCAUCUUGGCCAACUUGCAGCAG------GCAAUACGGGUCAAUUGCUGCGUUAAGUGCAUUGGCU ........(((((.(((..((((((((((((.....)))))....(((((((------((.......)))...))))))))))))))))))))). ( -33.80) >DroSim_CAF1 101595 89 + 1 UGCAAGCCGCUAACUGCCAACUUGGCUGGCCAUCUUGGCCAACUUGCAGCAG------GCAAUGCGGGUCAAUUGCUGCGUUAAGUGCAUUGGCU ........(((((.(((..((((((((((((.....)))))....(((((((------((.......)))...))))))))))))))))))))). ( -33.80) >DroEre_CAF1 101772 88 + 1 UGCGAGCCGCAGGCUGCCAACUUGGCUGGCCAUCUUGGCCAAC-UGCAGCAG------GCAAUGCGGGUCAAUUGCAGAGCUGAGUGCAUUGCCA .....((.((((...(((.....)))(((((.....))))).)-))).)).(------(((((((((.((.......)).))....)))))))). ( -37.70) >DroYak_CAF1 106476 71 + 1 ----AUCUGCAAGCUGCUAACUU-------------GGCCAAC-UGUAGUUG------GCAAUGCGGGUCAAUUGCAGAGUUGAGUGCAUUGGCA ----.(((((((...(((..(..-------------.((((((-....))))------))...)..)))...)))))))((..(.....)..)). ( -25.70) >consensus UGCAAGCCGCUAACUGCCAACUUGGCUGGCCAUCUUGGCCAAC_UGCAGCAG______GCAAUGCGGGUCAAUUGCUGCGUUAAGUGCAUUGGCA ........(((((.(((..((((((((((((.....))))).................(((((........)))))...))))))))))))))). (-20.66 = -22.10 + 1.44)



| Location | 17,104,101 – 17,104,195 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -18.36 |
| Energy contribution | -19.88 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17104101 94 - 22224390 GGCCAAUGCACUUAACGUAGCAAUUGACCUGCAUUGCGUUCAACUGCUGCA-GUUGGCCAAGAUGGCCAGCCAAGUUGGCAGUUAGCUGCUUACA (((.............((((((.((((..(((...))).)))).)))))).-..(((((.....))))))))..((.(((((....))))).)). ( -34.50) >DroSec_CAF1 110877 89 - 1 AGCCAAUGCACUUAACGCAGCAAUUGACCCGUAUUGC------CUGCUGCAAGUUGGCCAAGAUGGCCAGCCAAGUUGGCAGUUAGCGGCUUGCA ......(((.......)))((((.....(((((((((------(.(((....(((((((.....)))))))..))).))))))..)))).)))). ( -32.80) >DroSim_CAF1 101595 89 - 1 AGCCAAUGCACUUAACGCAGCAAUUGACCCGCAUUGC------CUGCUGCAAGUUGGCCAAGAUGGCCAGCCAAGUUGGCAGUUAGCGGCUUGCA ......(((.......)))((((.....(((((((((------(.(((....(((((((.....)))))))..))).))))))..)))).)))). ( -34.20) >DroEre_CAF1 101772 88 - 1 UGGCAAUGCACUCAGCUCUGCAAUUGACCCGCAUUGC------CUGCUGCA-GUUGGCCAAGAUGGCCAGCCAAGUUGGCAGCCUGCGGCUCGCA .((((((((..((((........))))...)))))))------).((((((-(.(((((.....)))))(((.....)))...)))))))..... ( -39.70) >DroYak_CAF1 106476 71 - 1 UGCCAAUGCACUCAACUCUGCAAUUGACCCGCAUUGC------CAACUACA-GUUGGCC-------------AAGUUAGCAGCUUGCAGAU---- (((....)))......((((((((((.(..((...((------((((....-)))))).-------------..))..)))).))))))).---- ( -20.40) >consensus AGCCAAUGCACUUAACGCAGCAAUUGACCCGCAUUGC______CUGCUGCA_GUUGGCCAAGAUGGCCAGCCAAGUUGGCAGUUAGCGGCUUGCA ...((((((..((((........))))...)))))).........(((((....(((((.....)))))(((.....))).....)))))..... (-18.36 = -19.88 + 1.52)

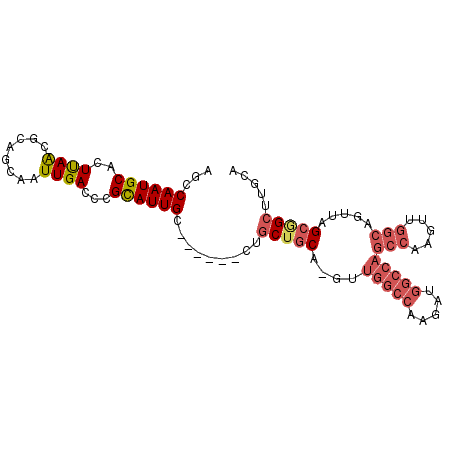

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:35 2006