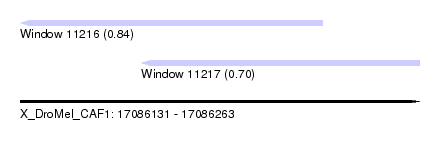
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,086,131 – 17,086,263 |
| Length | 132 |
| Max. P | 0.836183 |

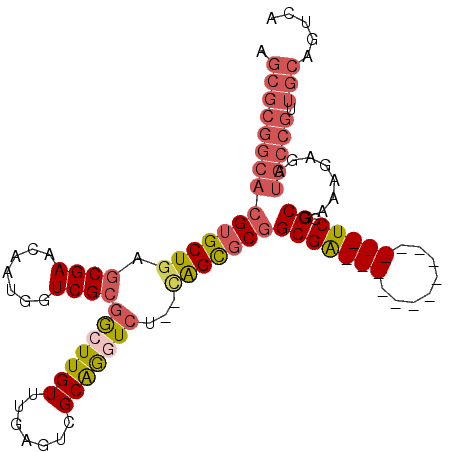
| Location | 17,086,131 – 17,086,231 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -36.61 |
| Consensus MFE | -28.36 |
| Energy contribution | -29.52 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17086131 100 - 22224390 GGCUUGUUUGAGUCGCAGGUCUCAGCGCGGCGA--------------UCGGCGAAAGAGAUGCCGUUGCAGUCAGCGAAAGAGAUGGCGGAAAGGCCGGAGAAAGAGACGGCGA (((((....)))))((..(((((.(((((((.(--------------((..(....).))))))).))).((((.(....)...))))((.....)).......))))).)).. ( -37.70) >DroSec_CAF1 92907 100 - 1 GGCUUGUUUGAGUCGCAGGUCUCAGCGCGGCGA--------------UCGGCGAAAAAGAUGCCGUUGCAGGCAGCGAAAGAGAUGGAGGAAAGGCCGGAGAAAGAGACGGCGA (((((....)))))((..(((((.(((((((.(--------------((.........))))))).))).(((..(....).............))).......))))).)).. ( -31.76) >DroSim_CAF1 83595 100 - 1 GGCUUGUUUGAGUCGCAAGUCUCAGCGCGGCGA--------------UCGGCGAAAGAGAUGCCGUUGCAGGCAGCGAAAGAGAUGGAGGAAAGGCCGGAGAAAGAGACGGCGA .(((((((((((.(....).))))).(((((.(--------------((..(....).)))))))).))))))..(....).............((((..........)))).. ( -35.40) >DroEre_CAF1 84432 100 - 1 GGCUUGUUUGAGUCGCAGCUCUUGGCGCGGCGA--------------UCGGCGAAAGAGAUGCCGUUGCAGUCGGCGAAAGAGAUGGCGGCCAGGCCGGAGAAAGAGACGGCGA (((((....))))).......((((((((((.(--------------((..(....).))))))))(((.(((..(....).))).))))))))((((..........)))).. ( -37.50) >DroYak_CAF1 88482 114 - 1 GACUUGUUUGAGUCGCAGGUCUUGGCGCGGCGAUCGUUGAUCGUUGAUCGGCGAAAGAGAUGCCGUUGCAGUCGGCGAAAGAGAUGGUGGCAAGGCCAGAGAAAGAGACGGCGA (((((....)))))((..((((((((.((((((((...))))))))(((..(....).))))))((..(.(((..(....).))).)..)).............))))).)).. ( -40.70) >consensus GGCUUGUUUGAGUCGCAGGUCUCAGCGCGGCGA______________UCGGCGAAAGAGAUGCCGUUGCAGUCAGCGAAAGAGAUGGAGGAAAGGCCGGAGAAAGAGACGGCGA (((((....)))))....(((((....((.((((((........)))))).))...)))))(((((..(.((((.(....)...))))((.....)).......)..))))).. (-28.36 = -29.52 + 1.16)

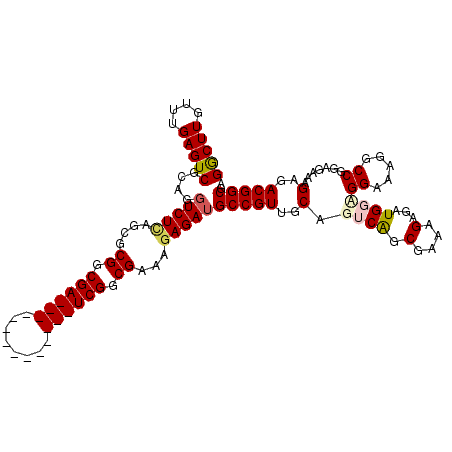
| Location | 17,086,171 – 17,086,263 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -38.13 |
| Consensus MFE | -25.35 |
| Energy contribution | -27.40 |
| Covariance contribution | 2.05 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17086171 92 - 22224390 AGCGCGGCACGUGCUGAGCGAACAAUGGUCGCGGCUUGUUUGAGUCGCAGGUCU--CAGCGCGGCGA--------------UCGGCGAAAGAGAUGCCGUUGCAGUCA .(((((((.((((((((((((.......))(((((((....)))))))..).))--)))))))...(--------------((..(....).))))))).)))..... ( -40.90) >DroGri_CAF1 100642 74 - 1 AGCGCGGCACGUGCUAUGCGAACAAUGGUCGACGCUUGUUUGAUUCGCGGCUCUUUCAGU-CUGCGG--------------UCUGCGAG------------------- ..(((((.((.(((...((((((((.(((....)))...))).)))))((((.....)))-).))))--------------))))))..------------------- ( -21.80) >DroSec_CAF1 92947 92 - 1 AGCGCGGCACGUGCUGAGCGAACAAUGGUCGCGGCUUGUUUGAGUCGCAGGUCU--CAGCGCGGCGA--------------UCGGCGAAAAAGAUGCCGUUGCAGGCA .((((((((((((((((((((.......))(((((((....)))))))..).))--)))))))((..--------------...))........))))).)))..... ( -39.40) >DroSim_CAF1 83635 92 - 1 AGCGCGGCACGUGCUGAGCGAACAAUGGUCGCGGCUUGUUUGAGUCGCAAGUCU--CAGCGCGGCGA--------------UCGGCGAAAGAGAUGCCGUUGCAGGCA .(((((((..(((((((((((.......))(((((((....)))))))..).))--))))))(((.(--------------((..(....).)))))))))))..)). ( -41.40) >DroEre_CAF1 84472 92 - 1 AGCGCGGCACGUGCUGAGCGAACAAUGGUCGCGGCUUGUUUGAGUCGCAGCUCU--UGGCGCGGCGA--------------UCGGCGAAAGAGAUGCCGUUGCAGUCG .(((((((.((((((((((((.......))(((((((....))))))).)))).--.))))))...(--------------((..(....).))))))).)))..... ( -39.50) >DroYak_CAF1 88522 106 - 1 AGCGCGGCACGUGCUGAGCGAACAAUGGUCGCGACUUGUUUGAGUCGCAGGUCU--UGGCGCGGCGAUCGUUGAUCGUUGAUCGGCGAAAGAGAUGCCGUUGCAGUCG .(((((((((.((((((((((.(((((((((((((((....)))))))..(((.--......))))))))))).))))...)))))).....).))))).)))..... ( -45.80) >consensus AGCGCGGCACGUGCUGAGCGAACAAUGGUCGCGGCUUGUUUGAGUCGCAGGUCU__CAGCGCGGCGA______________UCGGCGAAAGAGAUGCCGUUGCAGUCA .(((((((((((((((.((((.......))))(((((((.......)))))))...)))))))(((((((........))))).))........))))).)))..... (-25.35 = -27.40 + 2.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:28 2006