
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,084,450 – 17,084,553 |
| Length | 103 |
| Max. P | 0.874551 |

| Location | 17,084,450 – 17,084,553 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.88 |
| Mean single sequence MFE | -44.07 |
| Consensus MFE | -27.73 |
| Energy contribution | -28.40 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17084450 103 - 22224390 UUCCUGCUAAUCUGGCUGAUGCGGCUCAUCCGGCUGGCGUGCGAACCGCGCUCCGAGUUGCACUUGGAGCCGCAGAGCGAGUUCUCGCAGAAGCGACAGCACU ...((((....((.(((..((((((((...(((((((.(((((...))))).)).)))))......)))))))).))).)).....))))..((....))... ( -43.40) >DroVir_CAF1 104089 94 - 1 ------UUGGCGCGAUUG---CAGCUGAGGCGGCUCGCAUGGGAGCCGCGCUCCGAAUUGCAUUUGGAGCCGCAGAGUGAGUUGUCGCAGUAGCGGCAGCAUU ------....(((..(((---(.......(((((((......)))))))(((((((((...))))))))).)))).))).((((((((....))))))))... ( -45.40) >DroGri_CAF1 98650 93 - 1 ----------UGCGAUUGUUGCAGCUGAGGCGACUCGCAUGCGAGCUGCGCUCCGAGUUGCACUUGGAGCCGCACAUUGAGUUGUCGCAGAAGCGGCAACAUU ----------((((......(((((((((....)))(....).))))))((((((((.....))))))))))))......((((((((....))))))))... ( -45.00) >DroSim_CAF1 82018 103 - 1 UUCCUGCUAAUCUGGCUGAUGCGGCUCAUCCGGCUGGCGUGCGAACCGCGCUCCGAGUUGCACUUGGAGCCGCAGAGCGAGUUCUCGCAGAAGCGGCAGCACU .....((((..((((.(((......))).)))).))))((((...((((((((((((.....))))))))......((((....))))....))))..)))). ( -43.80) >DroMoj_CAF1 109478 103 - 1 UUGUUGUUGUUGCGAUUGCUGCAGCUGAGCCGACUCGCAUGGGAGCCGCGCUCCGAGUUGCACUUGGAGCCGCAGAGCGAAUUGUCGCAGAAGCGGCAGCACU ..(((((....)))))((((((.(((....((((((((..((...))((((((((((.....))))))).)))...))))...))))....))).)))))).. ( -42.50) >DroAna_CAF1 82352 103 - 1 CCCCGGCUUAUCUGGCUGAUCCGGCUGAGGCGACUGGCGUGGGAGCCGCGCUCCGAGUUGCACUUGGAGCCGCAGAGGGAGUUCUCGCAGAAGCGACAGCACU .(((((((((((.....)))((((((.((....))))).))))))))((((((((((.....))))))).)))...))).(((.((((....)))).)))... ( -44.30) >consensus UUCCUGCUAAUCCGACUGAUGCAGCUGAGCCGACUCGCAUGCGAGCCGCGCUCCGAGUUGCACUUGGAGCCGCAGAGCGAGUUCUCGCAGAAGCGGCAGCACU ...................((((((.(((....))))).)))).(((((((((((((.....))))))))......((((....))))....)))))...... (-27.73 = -28.40 + 0.67)


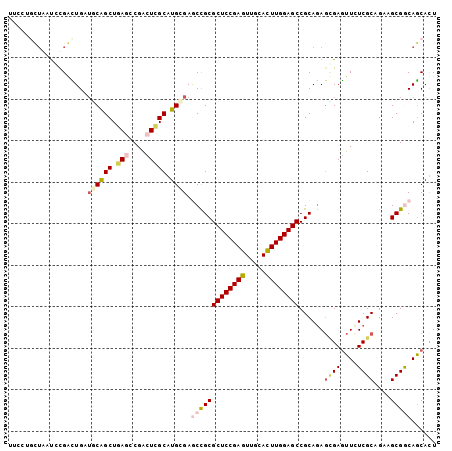
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:26 2006