
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,055,005 – 17,055,116 |
| Length | 111 |
| Max. P | 0.808924 |

| Location | 17,055,005 – 17,055,116 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -15.62 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
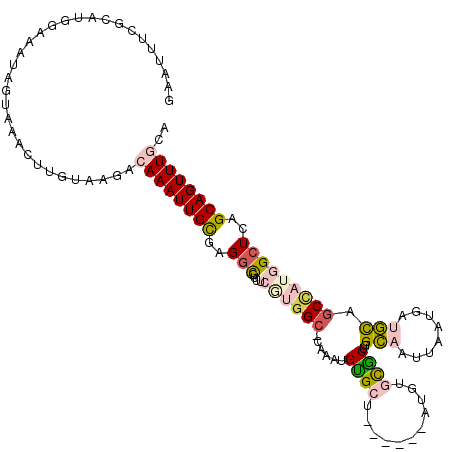
>X_DroMel_CAF1 17055005 111 + 22224390 GAAUUUCGCAUAGAAAUAGUAAACUUGUAAGACAAAUUGCUAGGCGUUUCGUGGC-CAAAUGUGCU------AUGUGCGCUGGCAAUUAAUGAUGCAGCCAUGGCUCAGCAGUUUGCA ..(((((.....))))).(((((((.((......(((((((((.(((..((((((-(....).)))------))).)))))))))))).....((.(((....)))))))))))))). ( -36.30) >DroVir_CAF1 75064 103 + 1 AAAUUU-------AAAUUGUAAA-UUGUAAGACAAAUUGUGAGCUGCU----GGC-AAAAAGUGUUGCCGCAAAA--UGCUUGUAAUUAAUGACAUCGCUAUGGCUCAGCAGUUUACA ......-------....((((((-((((......(((((..((((((.----(((-((......))))))))...--.)))..))))).........((....))...)))))))))) ( -27.70) >DroGri_CAF1 65264 104 + 1 GAAAUU-------AAAUUGUAAA-CAGUCAGACAAAUUGUGAGGCGCU----UGAAAAAAAGUGUUGCUGCAAAA--UACUUGUAAUUAAUGACAUCGCUAUGGCUCAGCAGUUUACA ......-------....((((((-(.((((....(((((..(((((((----(......))))))..........--..))..)))))..))))...(((.......))).))))))) ( -22.10) >DroSec_CAF1 54818 111 + 1 GAAUUUCGCAUGGAAAUAGUAAACUUGUAAGACAAAUUGCGAGGCGUUUCAUGGC-CAAAUGUGCU------AUGUGCGCUGGCAAUUAAUGAUGCAGCCAUGGCUCAGCAGUUUGCA ..(((((.....))))).(((((((.((......((((((..(((((..((((((-(....).)))------))).))))).)))))).....((.(((....)))))))))))))). ( -35.20) >DroEre_CAF1 55215 111 + 1 GAAUUUCGCAUGGAUAUAGUGAGCUUGUAAGACAAAUUGCGAGGCGUUUCGUGGC-CAAAUGUGCU------AUGUGCGCUGGCAAUUAAUGAUGCAGCCAUGGCUCAGCAGUUUGCA ..................(..((((.((......((((((..(((((..((((((-(....).)))------))).))))).)))))).....((.(((....)))))))))))..). ( -32.50) >DroYak_CAF1 57027 111 + 1 GAAUUUCGCAUGGAUAUAGUAAACUUGUAAGACAAAUUGCGAGGCGUUUCGUGGC-CAAAUGCGCC------AUGUGCGCUGGCAAUUAAUGAUGCAGCCAUGGCUCAGCAGUUUGCA ..................(((((((.((......((((((..(((((..((((((-(....).)))------))).))))).)))))).....((.(((....)))))))))))))). ( -35.30) >consensus GAAUUUCGCAUGGAAAUAGUAAACUUGUAAGACAAAUUGCGAGGCGUUUCGUGGC_CAAAUGUGCU______AUGUGCGCUGGCAAUUAAUGAUGCAGCCAUGGCUCAGCAGUUUGCA ................................((((((((..(((....((((((......((((...........))))..(((........))).)))))))))..)))))))).. (-15.62 = -16.40 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:21 2006