
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,044,916 – 17,045,016 |
| Length | 100 |
| Max. P | 0.970877 |

| Location | 17,044,916 – 17,045,016 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -17.17 |
| Energy contribution | -18.17 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
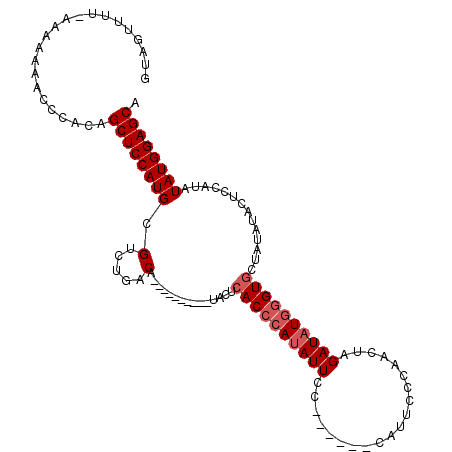
>X_DroMel_CAF1 17044916 100 + 22224390 GUAGUUUUUAAAAAAACCCACAGCUCCAUGCGUCUGACA----------UACUCACCAAUAUUCC------CAUUCCCAACAAGAUAUGGGUGCUAUAUACUCCAUAUAUGGAGCA ((.(((((....)))))..)).((((((((....(((..----------...)))...((((.((------(((((.......)).)))))....))))........)))))))). ( -19.00) >DroSec_CAF1 44607 99 + 1 GUAGUUUU-AAAAAAACCCACAGCUCCAUGCGUCUGACA----------UGCUCACCCAAAUUCC------CAUUCCCAACUAGAUAUGGGUGCUAUAUACUCCAUAUAUGGAGCG ((.((((.-....))))..)).(((((((((((.....)----------))).((((((.(((..------............))).))))))...............))))))). ( -21.64) >DroSim_CAF1 37870 99 + 1 GUAGUUUU-CAAAAAACCCACAGCUCCAUGCGUCUGACA----------UGCUCACCCAUAUUCC------CAUUCCCAACUAGAUAUGGGUGCUAUAUACUCCAUAUAUGGAGCG ((.((((.-....))))..)).(((((((((((.....)----------))).((((((((((..------............))))))))))...............))))))). ( -25.74) >DroYak_CAF1 46723 115 + 1 GUAUUUUC-CACAAAAUUCGCAGCUCCAUGCGUCUGACACACGACGACACACUCACCCAUAUUCCAAUUUCCAUUCCCAAACAGAUAUGAGUACUAUAUGCUCCAUAUAUGGAGCA ........-.............(((((((.((((........))))......................................(((((((((.....)))).)))))))))))). ( -21.10) >consensus GUAGUUUU_AAAAAAACCCACAGCUCCAUGCGUCUGACA__________UACUCACCCAUAUUCC______CAUUCCCAACUAGAUAUGGGUGCUAUAUACUCCAUAUAUGGAGCA ......................((((((((.(.....)...............((((((((((....................))))))))))..............)))))))). (-17.17 = -18.17 + 1.00)

| Location | 17,044,916 – 17,045,016 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -21.13 |
| Energy contribution | -20.75 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17044916 100 - 22224390 UGCUCCAUAUAUGGAGUAUAUAGCACCCAUAUCUUGUUGGGAAUG------GGAAUAUUGGUGAGUA----------UGUCAGACGCAUGGAGCUGUGGGUUUUUUUAAAAACUAC (((((((....)))))))((((((.(((((.(((....))).)))------))....((.(((.((.----------......)).))).))))))))((((((....)))))).. ( -28.60) >DroSec_CAF1 44607 99 - 1 CGCUCCAUAUAUGGAGUAUAUAGCACCCAUAUCUAGUUGGGAAUG------GGAAUUUGGGUGAGCA----------UGUCAGACGCAUGGAGCUGUGGGUUUUUUU-AAAACUAC .((((((....)))))).((((((.(((((.(((....))).)))------))............((----------(((.....)))))..))))))(((((....-.))))).. ( -27.80) >DroSim_CAF1 37870 99 - 1 CGCUCCAUAUAUGGAGUAUAUAGCACCCAUAUCUAGUUGGGAAUG------GGAAUAUGGGUGAGCA----------UGUCAGACGCAUGGAGCUGUGGGUUUUUUG-AAAACUAC .((((((....)))))).(((((((((((((((((........))------)..))))))))...((----------(((.....)))))..))))))(((((....-.))))).. ( -29.80) >DroYak_CAF1 46723 115 - 1 UGCUCCAUAUAUGGAGCAUAUAGUACUCAUAUCUGUUUGGGAAUGGAAAUUGGAAUAUGGGUGAGUGUGUCGUCGUGUGUCAGACGCAUGGAGCUGCGAAUUUUGUG-GAAAAUAC .((((((....))))))(((((.(((((((((((((((........))).))).)))))))))..)))))..(((((((.....)))))))..(..(((...)))..-)....... ( -28.70) >consensus CGCUCCAUAUAUGGAGUAUAUAGCACCCAUAUCUAGUUGGGAAUG______GGAAUAUGGGUGAGCA__________UGUCAGACGCAUGGAGCUGUGGGUUUUUUG_AAAACUAC .((((((....))))))......(((((((((((.................)).))))))))).....................((((......))))((((((....)))))).. (-21.13 = -20.75 + -0.37)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:18 2006