
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,025,689 – 17,025,790 |
| Length | 101 |
| Max. P | 0.940573 |

| Location | 17,025,689 – 17,025,790 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -18.51 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
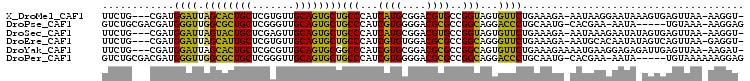
>X_DroMel_CAF1 17025689 101 + 22224390 -ACCUU-UUAACUCACUUUAUUCCUUAUU-UCUUUCAGAACACUACCGGCACGUCCGCAUGAUGGGCAGCACUGCAACACGAGCAGUGCUAAUCCAUCG---CAGAA -.....-.....................(-(((.............(((.....)))...((((((.((((((((.......))))))))..)))))).---.)))) ( -23.00) >DroPse_CAF1 38223 99 + 1 CUCCUU-UUUACA-----UAUU-UUCGUG-CAUUGCAGGGUCCUGCCGGCGCGUCCCCACGAUGGGCAGCACUGCAACCCGAGCAGCGCCAACCCAUCGUCGCAGAC ......-......-----....-...(((-(...((((....))))..))))(((..(((((((((..((.((((.......)))).))...)))))))).)..))) ( -36.70) >DroSec_CAF1 30468 101 + 1 -ACCUU-UUAACUCACUAUAUUCUUUAUU-UCUUUCAGAACACUACCGGCACGUCCGCAUGAUGGGCAGCACUGCAACUCGAGCAGUACUAAUCCAUCG---CAGAA -.....-.............((((.....-................(((.....)))...((((((.((.(((((.......))))).))..)))))).---.)))) ( -16.50) >DroEre_CAF1 30789 101 + 1 -ACCUC-UUAACUGACUAUAUUGUGCAUU-UCUUUCAGAACCCUGCCGGCGCGUCCACACGAUGGGCAGCACUGCAACACGAGCAAUGCUAAUCCAUCG---CAGAA -.....-....(((........((((...-.....(((....)))...)))).......(((((((.((((.(((.......))).))))..)))))))---))).. ( -24.50) >DroYak_CAF1 31867 102 + 1 -AUCUU-UUAACUCAAUCUCUCCUUCAUUUUCUUUCAGAACACUGCCGGCGCGUCCGCACGAUGGGCCGCACUGCAACGCGAGCAGUGCUAAUCCAUCG---CAGAA -.....-...................................((((.(((.((((.....)))).)))(((((((.......))))))).........)---))).. ( -29.00) >DroPer_CAF1 36290 100 + 1 CUCCUUUUUUACA-----UAUU-UUCGUG-CAUUGCAGGGUCCUGCCGGCGCGUCCCCACGAUGGGCAGCACUGCAACCCGAGCAGCGCCAACCCAUCGUCGCAGAC .............-----....-...(((-(...((((....))))..))))(((..(((((((((..((.((((.......)))).))...)))))))).)..))) ( -36.70) >consensus _ACCUU_UUAACUCACU_UAUUCUUCAUU_UCUUUCAGAACACUGCCGGCGCGUCCGCACGAUGGGCAGCACUGCAACACGAGCAGUGCUAAUCCAUCG___CAGAA ..........................................(((..((.....))...(((((((..(((((((.......)))))))...)))))))...))).. (-18.51 = -19.07 + 0.56)



| Location | 17,025,689 – 17,025,790 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -36.36 |
| Consensus MFE | -25.10 |
| Energy contribution | -23.05 |
| Covariance contribution | -2.05 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
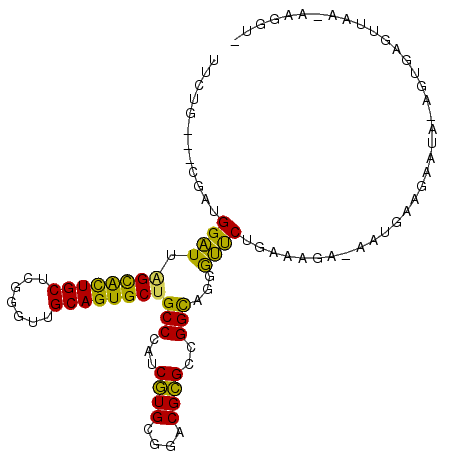
>X_DroMel_CAF1 17025689 101 - 22224390 UUCUG---CGAUGGAUUAGCACUGCUCGUGUUGCAGUGCUGCCCAUCAUGCGGACGUGCCGGUAGUGUUCUGAAAGA-AAUAAGGAAUAAAGUGAGUUAA-AAGGU- ..(((---((((((..(((((((((.......))))))))).)))))...(((.....)))))))((((((...(..-..)..))))))...........-.....- ( -31.90) >DroPse_CAF1 38223 99 - 1 GUCUGCGACGAUGGGUUGGCGCUGCUCGGGUUGCAGUGCUGCCCAUCGUGGGGACGCGCCGGCAGGACCCUGCAAUG-CACGAA-AAUA-----UGUAAA-AAGGAG ((((.(.(((((((((.((((((((.......)))))))))))))))))).))))(((...((((....))))..))-).....-....-----......-...... ( -45.80) >DroSec_CAF1 30468 101 - 1 UUCUG---CGAUGGAUUAGUACUGCUCGAGUUGCAGUGCUGCCCAUCAUGCGGACGUGCCGGUAGUGUUCUGAAAGA-AAUAAAGAAUAUAGUGAGUUAA-AAGGU- .((((---((((((..(((((((((.......))))))))).)))))..)))))...((((.((.((((((...(..-..)..)))))))).)).))...-.....- ( -30.00) >DroEre_CAF1 30789 101 - 1 UUCUG---CGAUGGAUUAGCAUUGCUCGUGUUGCAGUGCUGCCCAUCGUGUGGACGCGCCGGCAGGGUUCUGAAAGA-AAUGCACAAUAUAGUCAGUUAA-GAGGU- (((((---((((((..(((((((((.......))))))))).)))))))...(((......(((...(((.....))-).)))........))).....)-)))..- ( -32.34) >DroYak_CAF1 31867 102 - 1 UUCUG---CGAUGGAUUAGCACUGCUCGCGUUGCAGUGCGGCCCAUCGUGCGGACGCGCCGGCAGUGUUCUGAAAGAAAAUGAAGGAGAGAUUGAGUUAA-AAGAU- (((((---((((((....(((((((.......)))))))...)))))))..((((((.......))))))....))))......................-.....- ( -32.30) >DroPer_CAF1 36290 100 - 1 GUCUGCGACGAUGGGUUGGCGCUGCUCGGGUUGCAGUGCUGCCCAUCGUGGGGACGCGCCGGCAGGACCCUGCAAUG-CACGAA-AAUA-----UGUAAAAAAGGAG ((((.(.(((((((((.((((((((.......)))))))))))))))))).))))(((...((((....))))..))-).....-....-----............. ( -45.80) >consensus UUCUG___CGAUGGAUUAGCACUGCUCGGGUUGCAGUGCUGCCCAUCGUGCGGACGCGCCGGCAGGGUUCUGAAAGA_AAUGAAGAAUA_AGUGAGUUAA_AAGGU_ ............((((.((((((((.......))))))))(((...((((....))))..)))...))))..................................... (-25.10 = -23.05 + -2.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:14 2006