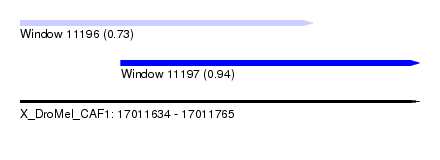
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,011,634 – 17,011,765 |
| Length | 131 |
| Max. P | 0.940495 |

| Location | 17,011,634 – 17,011,730 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -16.16 |
| Energy contribution | -15.83 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17011634 96 + 22224390 UUGGUCCCAGAAUAAGUGGAAAAAGGAAACAGCUAAUGAUGAUGAUUACCAUCCGACCAUCGUCAUUAUCAGGCGUCG---CCG----ACAUCGUCGUCAAUC- (((((.(((.......))).....(....).)))))((((((((((........(((....))).......(((...)---)).----..))))))))))...- ( -21.40) >DroPse_CAF1 22084 86 + 1 ----------------GGGAAAAAGGAAACUGUUAAUGAUGAUGAUUACCAUUC-AUCAUUAUCAUUAUCAGGCGCGGUCUCCGCUUUGCGCUGUCGUCAAUC- ----------------.............(((.(((((((((((((........-))))))))))))).)))((((((........))))))...........- ( -25.20) >DroSec_CAF1 16507 97 + 1 UUGGUCCCAGAAUAAGUGGAAAAAGGAAACAGCUAAUGAUGAUGAUUACCAUCCGACCAUCGUCAUUAUCAGGCGUCG---CCG----ACAUCGUCGUCAAUCC (((((.(((.......))).....(....).)))))((((((((((........(((....))).......(((...)---)).----..)))))))))).... ( -21.40) >DroEre_CAF1 17086 96 + 1 UAGGUCCCAGAAUAGCUGGAAAAAGGAAACUGCUAAUGAUGAUGAUUACCAUCCGACCAUCGUCAUUAUCAGGCGUCG---CCG----ACAUCGUCGUCAAUC- ..(((.((((.....))))......((..(((.(((((((((((.............))))))))))).)))...)))---))(----((......)))....- ( -27.62) >DroYak_CAF1 18149 96 + 1 UUGGUCCCAGAAUAAGUGGAAAAAGGAAACUGCUAAUGAUGAUGAUUACCAUCCGACCAUCGUCAUUAUCAGGCGUCG---CCG----ACAUCGUCGUCAAUC- ..(((.(((.......)))......((..(((.(((((((((((.............))))))))))).)))...)))---))(----((......)))....- ( -23.92) >DroPer_CAF1 19704 86 + 1 ----------------GGGAAAAAGGAAACUGUUAAUGAUGAUGAUUCCCAUUC-AUCAUUAUCAUUAUCAGGCGCGGUCUCCCCUUUGCGCUGUCGUCAAUC- ----------------.............(((.(((((((((((((........-))))))))))))).)))((((((........))))))...........- ( -25.20) >consensus UUGGUCCCAGAAUAAGUGGAAAAAGGAAACUGCUAAUGAUGAUGAUUACCAUCCGACCAUCGUCAUUAUCAGGCGUCG___CCG____ACAUCGUCGUCAAUC_ ........................(....).(.(((((((((((.............))))))))))).).((((.((..............)).))))..... (-16.16 = -15.83 + -0.33)



| Location | 17,011,667 – 17,011,765 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.57 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -14.30 |
| Energy contribution | -13.50 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17011667 98 + 22224390 UAAUGAUGAUGAUUACCAUCCGACCAUCGUCAUUAUCAGGCGUCG---CCG----ACAUCGUCGUCAAUC-GUUGGCAUGGCAUUCUGUGUCUAGUUCGCCU-----UGCC (((((((((((.............)))))))))))..(((((.((---(((----((.............-))))))..(((((...)))))..)..)))))-----.... ( -29.04) >DroPse_CAF1 22101 92 + 1 UAAUGAUGAUGAUUACCAUUC-AUCAUUAUCAUUAUCAGGCGCGGUCUCCGCUUUGCGCUGUCGUCAAUC-GUU--CUUGGCAU--UCC--------CCCCC-----UCCC (((((((((((((........-)))))))))))))...(((((((........)))))))...(((((..-...--.)))))..--...--------.....-----.... ( -25.60) >DroSec_CAF1 16540 97 + 1 UAAUGAUGAUGAUUACCAUCCGACCAUCGUCAUUAUCAGGCGUCG---CCG----ACAUCGUCGUCAAUCCGUU--CAUGGCAUUCUGUGUCUAGUUUGCCU-----UGUC (((((((((((.............)))))))))))...((((.((---...----....)).))))........--((.((((..(((....)))..)))).-----)).. ( -25.32) >DroEre_CAF1 17119 96 + 1 UAAUGAUGAUGAUUACCAUCCGACCAUCGUCAUUAUCAGGCGUCG---CCG----ACAUCGUCGUCAAUC-GUU--CAUGGCAUUCUGUGUCUAGUUUACCU-----UGCC (((((((((((.............)))))))))))...(((...)---))(----((((.(..((((...-...--..))))...).)))))..........-----.... ( -21.62) >DroYak_CAF1 18182 101 + 1 UAAUGAUGAUGAUUACCAUCCGACCAUCGUCAUUAUCAGGCGUCG---CCG----ACAUCGUCGUCAAUC-GUU--CAUGGUAUUCUGUGUCUAGUUUGCCUUGCCUUGCC (((((((((((.............)))))))))))...((((..(---(.(----((......)))....-...--...((((..(((....)))..))))..))..)))) ( -26.32) >DroPer_CAF1 19721 94 + 1 UAAUGAUGAUGAUUCCCAUUC-AUCAUUAUCAUUAUCAGGCGCGGUCUCCCCUUUGCGCUGUCGUCAAUC-GUU--CUUGGCAUUUUCU--------CCCCC-----CCCC (((((((((((((........-)))))))))))))...(((((((........)))))))...(((((..-...--.))))).......--------.....-----.... ( -25.60) >consensus UAAUGAUGAUGAUUACCAUCCGACCAUCGUCAUUAUCAGGCGUCG___CCG____ACAUCGUCGUCAAUC_GUU__CAUGGCAUUCUGUGUCUAGUUCGCCU_____UGCC (((((((((((.............)))))))))))...((((.................))))((((...........))))............................. (-14.30 = -13.50 + -0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:09 2006