
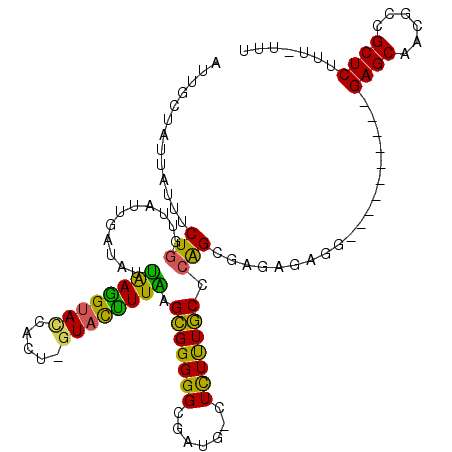
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,004,594 – 17,004,737 |
| Length | 143 |
| Max. P | 0.994680 |

| Location | 17,004,594 – 17,004,705 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 64.52 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -13.98 |
| Energy contribution | -12.66 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.39 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
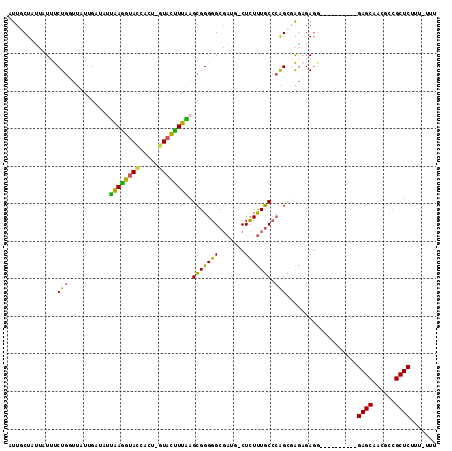
>X_DroMel_CAF1 17004594 111 + 22224390 UUUACUACUAUUUCUGUUUAUUGCCAUUAAGGUACCACU-GUACCUUAAGCGGGGGCGAUG-CUCUUUGCCCAGCUAGAGAGGGAGAGAGAGAGAGCAACGCCGCUCUUU-UUU ....((.((.((((((((.....((.(((((((((....-)))))))))..))((((((..-....))))))))).))))).)))).((((((((((......)))))))-))) ( -39.70) >DroSec_CAF1 9550 109 + 1 AUUGCUUUUAUAUCUGGUUACUGUUAUUAAGGUACCACU-GUACUUUAAGCGGGGGCGAUG-CUCUUUGCCCAGCUAGCGAGGGCGAAA--GAGAGCAACGCCGCUCUUU-UUU ...((((((....((((((.((((..(((((((((....-)))))))))))))((((((..-....)))))))))))).))))))((((--((((((......)))))))-))) ( -41.50) >DroSim_CAF1 4609 110 + 1 AUUGCUGGAAUGCCUGGAAAUUGA-AUAAAAGGACCACCCUUAUUUUUUGGGGGGGCCCUAACUUUCUUCCGGGGGGCCGAGGNNNNNN--NNNNNNNNNNNNNNNNNNN-NNN ....((.(..(.(((((((...((-(....(((.((.((((((.....)))))))).)))....)))))))))).)..).)).......--...................-... ( -28.50) >DroEre_CAF1 9342 102 + 1 AUUGCUAUUAUUUCUGGUUAGAGAUGCAAAGGUAUCACU-GUACUUUUAGCGGGGGCGAUG-CUCUUUGCGCAGUGAGAGAGG----------GAGCAACGCCGCUCUUUUUUU (((((....((((((....))))))(((((((((((.((-((.......))))....))))-).))))))))))).(((((((----------((((......))))))))))) ( -34.00) >DroYak_CAF1 10230 99 + 1 AUUACUAUUAUUUCUGCUUAAAGAUGUUGAGGUACCACU-GUACUUUGAGCGGGGGCGAUG-CUCUUUGCCCAGUGAGAGA------------GAGCAACACCGCUCUUU-UUC ...(((.......((((((((((((..((......))..-)).))))))))))((((((..-....)))))))))((((((------------((((......)))))))-))) ( -35.90) >consensus AUUGCUAUUAUUUCUGGUUAUUGAUAUUAAGGUACCACU_GUACUUUAAGCGGGGGCGAUG_CUCUUUGCCCAGCGAGAGAGG__________GAGCAACGCCGCUCUUU_UUU .............(((...........((((((((.....)))))))).(((((((.......))))))).)))...................((((......))))....... (-13.98 = -12.66 + -1.32)

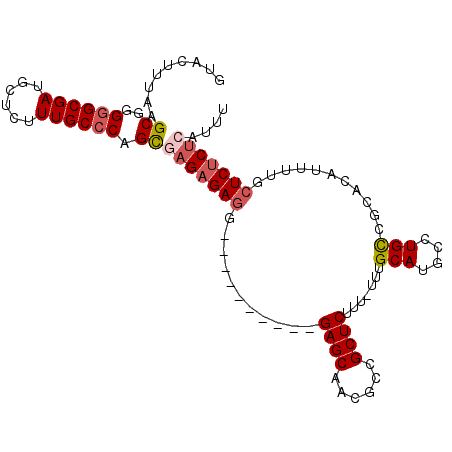
| Location | 17,004,633 – 17,004,737 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -21.88 |
| Energy contribution | -22.88 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
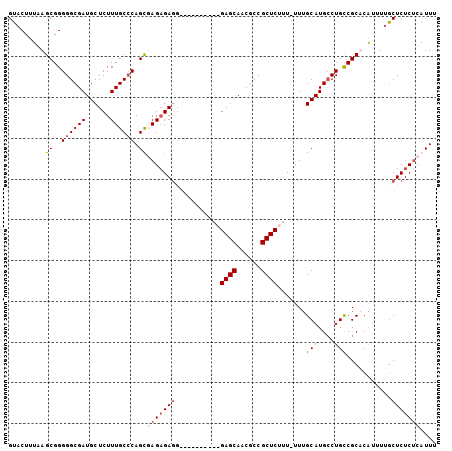
>X_DroMel_CAF1 17004633 104 + 22224390 GUACCUUAAGCGGGGGCGAUGCUCUUUGCCCAGCUAGAGAGGGAGAGAGAGAGAGCAACGCCGCUCUUU-UUUGCAUGCCUGUCGCACAUUUUGCUCUCGUAUUU ((((....(((..((((((......)))))).))).(((((.((((((((((((((......)))))))-)))...(((.....)))..)))).))))))))).. ( -38.90) >DroSec_CAF1 9589 102 + 1 GUACUUUAAGCGGGGGCGAUGCUCUUUGCCCAGCUAGCGAGGGCGAAA--GAGAGCAACGCCGCUCUUU-UUUGCAUGCCUGUCGCACAUUUUGCUCUCUUAUUU ........(((..((((((......)))))).)))(((((((((((((--((((((......)))))))-))))).(((.....)))..)))))))......... ( -37.80) >DroEre_CAF1 9381 94 + 1 GUACUUUUAGCGGGGGCGAUGCUCUUUGCGCAGUGAGAGAGG----------GAGCAACGCCGCUCUUUUUUUGCAUGGCUGCCGCUCAUUUGACUCUCUCAUU- ((......(((((.(((.((((.....))((((.(((((((.----------(.((...))).))))))).)))))).))).)))))......)).........- ( -32.70) >DroYak_CAF1 10269 92 + 1 GUACUUUGAGCGGGGGCGAUGCUCUUUGCCCAGUGAGAGA------------GAGCAACACCGCUCUUU-UUCGCAUGCCUGCCGCUCGUUUUGCUCUCUCAUUU (((...(((((((((((((......)))))).((((((((------------((((......)))))))-))))).......)))))))...))).......... ( -42.10) >consensus GUACUUUAAGCGGGGGCGAUGCUCUUUGCCCAGCGAGAGAGG__________GAGCAACGCCGCUCUUU_UUUGCAUGCCUGCCGCACAUUUUGCUCUCUCAUUU .........((..((((((......)))))).))(((((((...........((((......)))).......(((....)))...........))))))).... (-21.88 = -22.88 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:07 2006