
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,998,995 – 16,999,143 |
| Length | 148 |
| Max. P | 0.963846 |

| Location | 16,998,995 – 16,999,107 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -31.95 |
| Energy contribution | -31.57 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
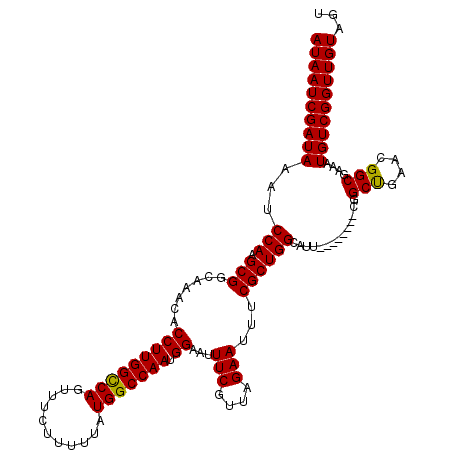
>X_DroMel_CAF1 16998995 112 + 22224390 AUAAUCGAUAAAUCCAAGCGUCAAACACCUUGGUCAGUUUCUUUUUAUGGCCAAUGGAAUUUCGUUAGAAUUUCGCUGGCAUU--------CGGCUGAACGGCGAAAUGUCGGUUGUAGC (((((((((....(((((.((.....)))))))...(((((........((((.((((((((.....)))))))).))))...--------..(((....)))))))))))))))))... ( -31.30) >DroSec_CAF1 4121 112 + 1 AUAAUCGAUAAAUCCAAGCGGCAAACACCUUGGCCAGUUUCUUUUUAUGGCCAAUGGAAUUUCGUUAGAAUUUCGCUGGCAUU--------CGGCCGAACGGCGAAAUGUCGGUUGUAGU ((((((((((...(((.((((......(((((((((...........))))))).))...(((....)))..)))))))..((--------((.((....)))))).))))))))))... ( -35.30) >DroEre_CAF1 3945 119 + 1 AUAAUCGAUAAAUCCAAGCGGCAAACACCUUGGCCAGUUUCUUUUUAUGGCCAAUGGAAUUUCGUUAGAAUUUCGCUGGCAUUCGCUGGCUUGGCUGAACGGCGAAAUGUCGGUUGUAG- ((((((((((..((((((((((.....(((((((((...........))))))).))......(((((.......)))))....))).)))))(((....)))))..))))))))))..- ( -39.40) >DroYak_CAF1 3946 112 + 1 AUAAUCGAUAAAUCCAAGCGGCAAACACCUUGGCCAGUUUCUUUUUAUGGCCAAUGGAAUUUCGUUAGAAUUUCGCUGGCAUU--------UGGCUCAACGGCGAAAUGUCGGUUGUACU ((((((..........(((((......(((((((((...........))))))).))...(((....)))..)))))((((((--------(.(((....))).)))))))))))))... ( -34.00) >consensus AUAAUCGAUAAAUCCAAGCGGCAAACACCUUGGCCAGUUUCUUUUUAUGGCCAAUGGAAUUUCGUUAGAAUUUCGCUGGCAUU________CGGCUGAACGGCGAAAUGUCGGUUGUAGU ((((((((((...(((.(((.......(((((((((...........))))))).))...(((....)))...))))))..............(((....)))....))))))))))... (-31.95 = -31.57 + -0.38)


| Location | 16,999,035 – 16,999,143 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -27.91 |
| Energy contribution | -27.47 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16999035 108 + 22224390 CUUUUUAUGGCCAAUGGAAUUUCGUUAGAAUUUCGCUGGCAUU--------CGGCUGAACGGCGAAAUGUCGGUUGUAGCUAUG-GACUGAAGGUCGGAAAUGUCCGGAAUUCCAUA .............(((((((((((.....((((((((((((((--------..(((....)))..)))))))))..........-(((.....))).)))))...))))))))))). ( -33.30) >DroSec_CAF1 4161 109 + 1 CUUUUUAUGGCCAAUGGAAUUUCGUUAGAAUUUCGCUGGCAUU--------CGGCCGAACGGCGAAAUGUCGGUUGUAGUUAUGGGACUGAAGGUCGGAAAUGUCCGGAAUUCCAUA .............(((((((((((.....((((((((((((((--------..(((....)))..)))))))))...........(((.....))).)))))...))))))))))). ( -35.30) >DroEre_CAF1 3985 115 + 1 CUUUUUAUGGCCAAUGGAAUUUCGUUAGAAUUUCGCUGGCAUUCGCUGGCUUGGCUGAACGGCGAAAUGUCGGUUGUAG-UAUG-GACUGAAGGUCGGAAAUGCCCGGAAUUCCAUA .............(((((((((((.....(((((((((((((((((((...........))))).))))))))).....-....-(((.....))).)))))...))))))))))). ( -37.20) >DroYak_CAF1 3986 108 + 1 CUUUUUAUGGCCAAUGGAAUUUCGUUAGAAUUUCGCUGGCAUU--------UGGCUCAACGGCGAAAUGUCGGUUGUACUUAUG-GACUGAAGGUCGGAAGUGCCAGGAAUUUCAUG .............((((((((((...........(((((((((--------(.(((....))).)))))))))).((((((.(.-(((.....))).)))))))..)))))))))). ( -34.20) >consensus CUUUUUAUGGCCAAUGGAAUUUCGUUAGAAUUUCGCUGGCAUU________CGGCUGAACGGCGAAAUGUCGGUUGUAGUUAUG_GACUGAAGGUCGGAAAUGCCCGGAAUUCCAUA .............(((((((((((((((.......)))))............(((....((((......((((((.((....)).))))))..)))).....))).)))))))))). (-27.91 = -27.47 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:03 2006