
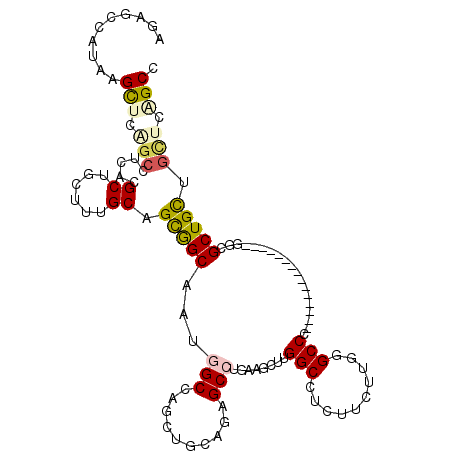
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,990,976 – 16,991,075 |
| Length | 99 |
| Max. P | 0.969243 |

| Location | 16,990,976 – 16,991,075 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 69.35 |
| Mean single sequence MFE | -42.57 |
| Consensus MFE | -16.27 |
| Energy contribution | -16.42 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16990976 99 + 22224390 AUAGCCAUUAGCUCAGCUUCCCGCUUCUUUGCAGCGGCAAUGGCUACUUGGAGGGCCUGAUGCUUGGCCUCUUCUUGUGCCC---------------GUCGCUGCUGUGUAGCC ..........(((((((.....))).....((((((((.((((.(((..((((((((........)))).))))..))).))---------------)).))))))))).))). ( -39.10) >DroPse_CAF1 2144 114 + 1 AGAGCCAUAAGUUCGGCCUCACGCUGCUUUGCCGCAGCUAUGGCCUGCUGCAGAGCCAUCAUUCUGGCCUCUUCCUGGGCCCUUUGGGCCUGCUGCUGGCGCUGUUGCUCUGCC (((((....(((..((((....(((((......)))))...)))).(((((((.((((......))))........((((((...)))))).)))).))))))...)))))... ( -48.20) >DroEre_CAF1 2069 99 + 1 AUGGCCAUGAGCUCAGCUUCUCGCUUCUUUGCAGCGGCAAUGGCUAUUUGGAGUGCCUGAUGCUUGGCCUCUUCUUGUGCCC---------------GGCGCUGCUGCUCAGCC ..(((..(((((..(((.....))).....((((((.(...(((.....((((.(((........)))))))......))).---------------).)))))).)))))))) ( -38.70) >DroYak_CAF1 2067 99 + 1 AUAGCCAUGAGUUCAGCUUCCCGCUUCUUCGCAGCGGCAAGUGCUAUUUGCAGGGCCUGAUGCUUGGCCUCUUCUUGUGCCC---------------GUCGCUGCUGCUCGGCC ...(((..((((..(((.....))).....((((((((...(((.....)))(((((........)))))............---------------)))))))).))))))). ( -35.40) >DroMoj_CAF1 2279 105 + 1 AGCUCCAGCAGCUCCUGCUCACGCUGCUUGGCCGUGGCUAUGGCCUGCUGCAGUGCAGCAAAUUUGGCCUCAUCGGCGGCCUUC---------UGCUGUCGCUGUUGCUCGGCA ....(.((((((..........)))))).)((((.(((....))).((.(((((((((((.....((((........))))...---------))))).)))))).)).)))). ( -45.20) >DroPer_CAF1 1352 114 + 1 AGAGCCAUAAGUUCGGCCUCACGCUGCUUGGCUGCAGCUAUGGCCUGCUGCAGAGCCAUCAUUCUGGCCUCUUCCUGGGCCCUUUGGGCCUGCUGCUGGCGCUGUUGCUCUGCC (((((....(((..((((....(((((......)))))...)))).(((((((.((((......))))........((((((...)))))).)))).))))))...)))))... ( -48.80) >consensus AGAGCCAUAAGCUCAGCCUCACGCUGCUUUGCAGCGGCAAUGGCCAGCUGCAGAGCCUGAAGCUUGGCCUCUUCUUGGGCCC_______________GGCGCUGCUGCUCAGCC ..........(((.(((.....((......)).(((((...(((..........)))........(((..........)))...................))))).))).))). (-16.27 = -16.42 + 0.14)

| Location | 16,990,976 – 16,991,075 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.35 |
| Mean single sequence MFE | -41.07 |
| Consensus MFE | -13.06 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16990976 99 - 22224390 GGCUACACAGCAGCGAC---------------GGGCACAAGAAGAGGCCAAGCAUCAGGCCCUCCAAGUAGCCAUUGCCGCUGCAAAGAAGCGGGAAGCUGAGCUAAUGGCUAU (((((.....((((...---------------.(((((..(.((.((((........)))))).)..)).))).((.(((((.......))))).))))))......))))).. ( -34.20) >DroPse_CAF1 2144 114 - 1 GGCAGAGCAACAGCGCCAGCAGCAGGCCCAAAGGGCCCAGGAAGAGGCCAGAAUGAUGGCUCUGCAGCAGGCCAUAGCUGCGGCAAAGCAGCGUGAGGCCGAACUUAUGGCUCU ...(((((......((..((((..(((((...)))))........(((((......))))))))).)).((((...(((((......)))))....)))).........))))) ( -46.90) >DroEre_CAF1 2069 99 - 1 GGCUGAGCAGCAGCGCC---------------GGGCACAAGAAGAGGCCAAGCAUCAGGCACUCCAAAUAGCCAUUGCCGCUGCAAAGAAGCGAGAAGCUGAGCUCAUGGCCAU ((((((((.(((((((.---------------.(((.......((((((........))).)))......)))...).)))))).....(((.....)))..))))..)))).. ( -38.72) >DroYak_CAF1 2067 99 - 1 GGCCGAGCAGCAGCGAC---------------GGGCACAAGAAGAGGCCAAGCAUCAGGCCCUGCAAAUAGCACUUGCCGCUGCGAAGAAGCGGGAAGCUGAACUCAUGGCUAU (((((((...((((..(---------------.((((.....((.((((........))))))((.....))...)))).((((......)))))..))))..)))..)))).. ( -34.70) >DroMoj_CAF1 2279 105 - 1 UGCCGAGCAACAGCGACAGCA---------GAAGGCCGCCGAUGAGGCCAAAUUUGCUGCACUGCAGCAGGCCAUAGCCACGGCCAAGCAGCGUGAGCAGGAGCUGCUGGAGCU (((...)))..(((..(((((---------(..(((((....((.((((.....((((((...)))))))))).))....))))).....((....)).....))))))..))) ( -45.70) >DroPer_CAF1 1352 114 - 1 GGCAGAGCAACAGCGCCAGCAGCAGGCCCAAAGGGCCCAGGAAGAGGCCAGAAUGAUGGCUCUGCAGCAGGCCAUAGCUGCAGCCAAGCAGCGUGAGGCCGAACUUAUGGCUCU ...(((((......((..((((..(((((...)))))........(((((......))))))))).)).((((...(((((......)))))....)))).........))))) ( -46.20) >consensus GGCAGAGCAACAGCGAC_______________GGGCACAAGAAGAGGCCAAACAUCAGGCACUGCAACAAGCCAUAGCCGCUGCAAAGAAGCGUGAAGCUGAACUCAUGGCUAU (((((............................(((.........((((........)))).........)))...((..((((......)).))..))........))))).. (-13.06 = -13.20 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:01 2006