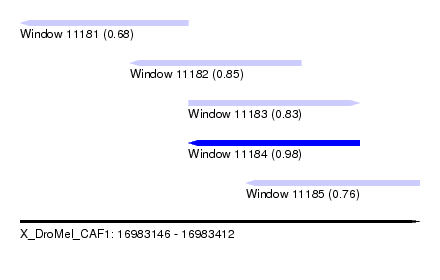
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,983,146 – 16,983,412 |
| Length | 266 |
| Max. P | 0.984853 |

| Location | 16,983,146 – 16,983,258 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.10 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -32.01 |
| Energy contribution | -32.90 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16983146 112 - 22224390 AAGCGGCGGAAGU-GAAAUUCUGUCCAUGAGAUAAAGCCAGAUCAUCCGAUUGAUUGGUCGGUGAUUACG------AAAUGAAUUUCCUGCCGGCGCUUUAUCCAAUUUCCCC-CGAAUU (((((.(((.((.-(((((((((((.....))))......(((((.((((((....)))))))))))...------....))))))))).))).)))))..............-...... ( -33.40) >DroEre_CAF1 11082 113 - 1 AAGCGGCGGCAGUCGAAAUGCUGUCCAUAAGAUAAAGCCAGAUCAUCCGAUUGAUGGGUCGGUGAUUACG------AAAUGAGUUUCCUGCCGGCGCUUUCUCCAUUUUCCAC-CGAAUU (((((.((((((..(((((((((((.....)))..)))..(((((.(((((......))))))))))...------......))))))))))).)))))..............-...... ( -34.70) >DroYak_CAF1 11588 120 - 1 AAGCGGCGGCAGUCGAAAUGCUGUCCAUAAGUUAAAGCCAGAUCAUCCGAUUGAUUGGUCGGUGAUUUCGUAAUCGUAAUGCGUUUGCUGCCGGCGCUUUCUCCAAUUUCCCCACGAAUU (((((.(((((((..((((((.........((((..((.((((((.((((((....)))))))))))).)).....))))))))))))))))).)))))..................... ( -37.20) >consensus AAGCGGCGGCAGUCGAAAUGCUGUCCAUAAGAUAAAGCCAGAUCAUCCGAUUGAUUGGUCGGUGAUUACG______AAAUGAGUUUCCUGCCGGCGCUUUCUCCAAUUUCCCC_CGAAUU (((((.((((((..(((((((((((.....))))......(((((.((((((....))))))))))).............))))))))))))).)))))..................... (-32.01 = -32.90 + 0.89)

| Location | 16,983,219 – 16,983,333 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.17 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -30.11 |
| Energy contribution | -29.67 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16983219 114 - 22224390 UAAAACA-UCCCAUGUUAUUGGGAAUCCUCUUAUUU--UUAAGGUUACGUAACCUUAACCCAUUUCGGGGGCCGGAA--AAAGCGGCGGAAGU-GAAAUUCUGUCCAUGAGAUAAAGCCA .......-(((((......)))))....((((((..--((((((((....))))))))(((......)))((((...--....))))(((...-(.....)..)))))))))........ ( -30.40) >DroEre_CAF1 11155 118 - 1 UGAAACA-UCCCAUGUUAUUGGGACUCCUCUUAUUU-UUUAAGGUUACGUAACCUUAACCCACUUCGGGGACCGGAAAAAAAGCGGCGGCAGUCGAAAUGCUGUCCAUAAGAUAAAGCCA .......-(((((......))))).(((........-.((((((((....))))))))(((.....)))....)))........(((((((((......))))))(....).....))). ( -30.80) >DroYak_CAF1 11668 118 - 1 UGAAAAAUUCCCAUGUCAUUGGGACUCCUCUUAUUUUUUUAAGGUUACGUAACCUUAACCCAUUUCAGGGUCCGGAA--AAAGCGGCGGCAGUCGAAAUGCUGUCCAUAAGUUAAAGCCA ........(((((......))))).(((...........(((((((....)))))))((((......))))..))).--...(.(((((((.......)))))))).............. ( -30.50) >consensus UGAAACA_UCCCAUGUUAUUGGGACUCCUCUUAUUU_UUUAAGGUUACGUAACCUUAACCCAUUUCGGGGACCGGAA__AAAGCGGCGGCAGUCGAAAUGCUGUCCAUAAGAUAAAGCCA ........(((((......))))).(((..........((((((((....))))))))(((......)))...)))......(.(((((((.......)))))))).............. (-30.11 = -29.67 + -0.44)


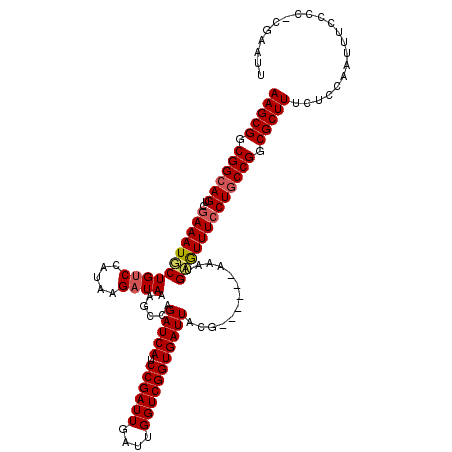
| Location | 16,983,258 – 16,983,372 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.89 |
| Mean single sequence MFE | -38.13 |
| Consensus MFE | -29.86 |
| Energy contribution | -30.64 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16983258 114 + 22224390 U--UUCCGGCCCCCGAAAUGGGUUAAGGUUACGUAACCUUAA--AAAUAAGAGGAUUCCCAAUAACAUGGGA-UGUUUUAUUGGUGCUGUCCUCCUAGCGGAAGAC-CCGCUAAAUCUGG .--..((((..(((.....)))((((((((....))))))))--......((((((.((((((((.((....-.)).))))))).)..)))))).((((((.....-))))))...)))) ( -35.20) >DroEre_CAF1 11195 118 + 1 UUUUUCCGGUCCCCGAAGUGGGUUAAGGUUACGUAACCUUAAA-AAAUAAGAGGAGUCCCAAUAACAUGGGA-UGUUUCAGUGGUGCUGUCCUCUUAGCGGGUGUCCCCGCUAAAACUGG .....(((((.(((.....)))((((((((....)))))))).-...((((((((((((((......)))))-)....(((.....)))))))))))(((((....)))))....))))) ( -43.20) >DroYak_CAF1 11708 117 + 1 U--UUCCGGACCCUGAAAUGGGUUAAGGUUACGUAACCUUAAAAAAAUAAGAGGAGUCCCAAUGACAUGGGAAUUUUUCAUUGGUGCUGUCCUCUCAGCGAAUGUC-CCGCUAAAUCUGG .--..((((((((......)))((((((((....)))))))).......(((((((..(((((((.(........).)))))))..)..)))))).((((......-.))))...))))) ( -36.00) >consensus U__UUCCGGACCCCGAAAUGGGUUAAGGUUACGUAACCUUAAA_AAAUAAGAGGAGUCCCAAUAACAUGGGA_UGUUUCAUUGGUGCUGUCCUCUUAGCGGAUGUC_CCGCUAAAUCUGG .....((((((((......)))((((((((....))))))))........(((((.(((((......))))).................))))).((((((......))))))..))))) (-29.86 = -30.64 + 0.78)

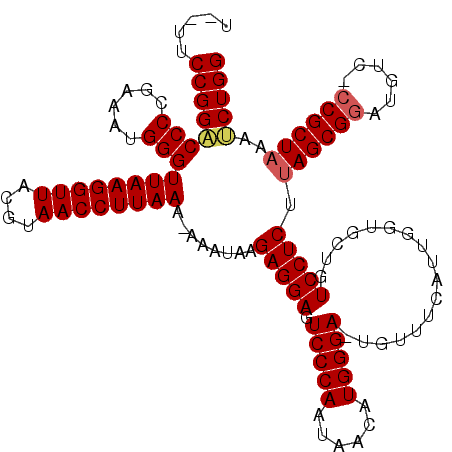

| Location | 16,983,258 – 16,983,372 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.89 |
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -33.80 |
| Energy contribution | -33.03 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16983258 114 - 22224390 CCAGAUUUAGCGG-GUCUUCCGCUAGGAGGACAGCACCAAUAAAACA-UCCCAUGUUAUUGGGAAUCCUCUUAUUU--UUAAGGUUACGUAACCUUAACCCAUUUCGGGGGCCGGAA--A .........((..-(((((((....))))))).))............-(((((......))))).(((........--((((((((....))))))))(((......)))...))).--. ( -36.20) >DroEre_CAF1 11195 118 - 1 CCAGUUUUAGCGGGGACACCCGCUAAGAGGACAGCACCACUGAAACA-UCCCAUGUUAUUGGGACUCCUCUUAUUU-UUUAAGGUUACGUAACCUUAACCCACUUCGGGGACCGGAAAAA ((.(((((((((((....))))))))((((((((.....))).....-(((((......))))).)))))......-.((((((((....))))))))(((.....)))))).))..... ( -42.60) >DroYak_CAF1 11708 117 - 1 CCAGAUUUAGCGG-GACAUUCGCUGAGAGGACAGCACCAAUGAAAAAUUCCCAUGUCAUUGGGACUCCUCUUAUUUUUUUAAGGUUACGUAACCUUAACCCAUUUCAGGGUCCGGAA--A ((......(((((-.....)))))(((((((...(.(((((((...((....)).))))))))..))))))).......(((((((....)))))))((((......))))..))..--. ( -33.30) >consensus CCAGAUUUAGCGG_GACAUCCGCUAAGAGGACAGCACCAAUGAAACA_UCCCAUGUUAUUGGGACUCCUCUUAUUU_UUUAAGGUUACGUAACCUUAACCCAUUUCGGGGACCGGAA__A ((....(((((((......)))))))(((((...(.(((((((............))))))))..)))))........((((((((....))))))))(((......)))...))..... (-33.80 = -33.03 + -0.77)

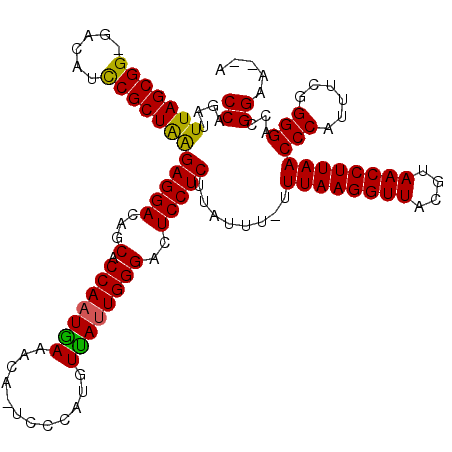
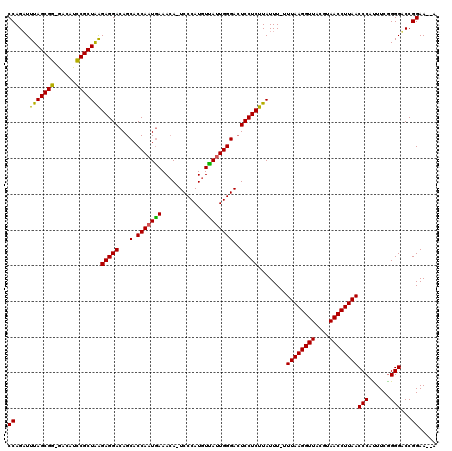
| Location | 16,983,296 – 16,983,412 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -25.39 |
| Energy contribution | -24.40 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16983296 116 - 22224390 AAAAUGGAUUUUAUGAGCACUUUCGUAAUUGAAUAAGUAGCCAGAUUUAGCGG-GUCUUCCGCUAGGAGGACAGCACCAAUAAAACA-UCCCAUGUUAUUGGGAAUCCUCUUAUUU--UU .....((((((..((.(((((((((....)))..)))).))))......((..-(((((((....))))))).)).((((((..(((-(...))))))))))))))))........--.. ( -30.20) >DroEre_CAF1 11235 117 - 1 AGUCUGGUGUU-UUGAGCACUUCCGUAAUUGAAAAAGUAGCCAGUUUUAGCGGGGACACCCGCUAAGAGGACAGCACCACUGAAACA-UCCCAUGUUAUUGGGACUCCUCUUAUUU-UUU ...(((((.((-(..(..((....))..)..))).....)))))..((((((((....))))))))((((((((.....))).....-(((((......))))).)))))......-... ( -38.20) >DroYak_CAF1 11746 119 - 1 AAACUUGUGUUUUUGAGCACUUCCGGAAUCGAAAAAGUAGCCAGAUUUAGCGG-GACAUUCGCUGAGAGGACAGCACCAAUGAAAAAUUCCCAUGUCAUUGGGACUCCUCUUAUUUUUUU ......(((((....)))))..........((((((((((......(((((((-.....)))))))(((((...(.(((((((...((....)).))))))))..))))))))))))))) ( -29.00) >consensus AAACUGGUGUUUUUGAGCACUUCCGUAAUUGAAAAAGUAGCCAGAUUUAGCGG_GACAUCCGCUAAGAGGACAGCACCAAUGAAACA_UCCCAUGUUAUUGGGACUCCUCUUAUUU_UUU .............((.((((((.((....))...)))).))))...(((((((......)))))))(((((...(.(((((((............))))))))..))))).......... (-25.39 = -24.40 + -0.99)

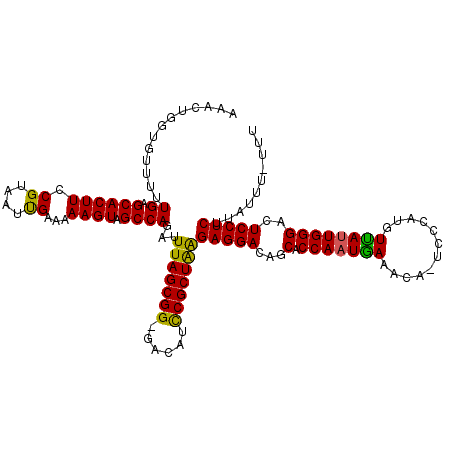
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:59 2006