| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,979,518 – 16,979,821 |
| Length | 303 |
| Max. P | 0.996507 |

| Location | 16,979,518 – 16,979,623 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.13 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -21.80 |
| Energy contribution | -20.44 |
| Covariance contribution | -1.36 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16979518 105 - 22224390 CUAAAACUGCAGCGUUGAUCCUUACAGGGUUGUGGUAUUCAACUGGACCAUAAUAAACCCCAUGAUUAGCUAUUCCUCUUAAGGGCAUGGCCUCAAAAAUGC-AAC .......((((...((((........((((((((((..........))))))....))))........(((((((((....)))).))))).))))...)))-).. ( -24.70) >DroVir_CAF1 6501 106 - 1 CUAAAACCGCAUCAGUGAUUUACACGCUGGCGUGGUAUUCAAUUGGACUACAUAAAAGCCCAUGAUUAACUAUUCCUCUUAAGGGCAUUGGCUCGAAGAUGCAAAC ........(((((.(((.....)))((..(.(((((..........)))))......((((.(((.............))).)))).)..)).....))))).... ( -24.32) >DroGri_CAF1 5680 106 - 1 CUAAAACCGCAUCAGUGAUUCACACGCUGAUGUGGUAUUCAAUUGGACUACAUCAAAGCCCAUGAUUUACUAUUCCUCUUAAGGGCAUUGGCUAAAAGAUGCGAAC .......((((((.(((.....)))(((((((((((..........))))))))...((((.(((.............))).))))...))).....))))))... ( -30.12) >DroSim_CAF1 6453 105 - 1 CUAAAACUGCAGCGUUGAUCUCUACAGCGUUGUGGUAUUCAACUGGACCACAAUAAACCCCAUGAUUAGCUAUUCCUCUUAAGGGCAUGGCCUCAAAAAUGA-AAC .....((..((((((((.......))))))))..)).((((..(((.............)))(((...(((((((((....)))).))))).)))....)))-).. ( -25.72) >DroMoj_CAF1 5860 106 - 1 CUAAAACCGCAUCAGUGACCCACACGCUGGUGUGGUAUUCAAUUGGACUACAUUGAACCCCAUGGUUAACUAUUCCUCUUAAGGGCAUUGGCUAAAAGAUGCAUUC ........(((((.(((.....)))((..((((((..((((((.(.....)))))))..)).............(((....)))))))..)).....))))).... ( -28.70) >DroAna_CAF1 9496 106 - 1 CUAAAACUGCAGCGUUGAUCCUCACAGCGUUGUGGUAUUCAAUUGGACCAAAAUGAACCCCAUGGUUAACUAUUCCUCUUAAGGGCAUUGCCUCAAAAAUGCGAAC .....((..((((((((.......))))))))..)).(((....(((........((((....))))......)))........(((((........)))))))). ( -25.44) >consensus CUAAAACCGCAGCAGUGAUCCACACACUGUUGUGGUAUUCAAUUGGACCACAAUAAACCCCAUGAUUAACUAUUCCUCUUAAGGGCAUUGCCUCAAAAAUGC_AAC .....((((((((((((.......))))))))))))..(((..(((.............)))))).........(((....)))(((((........))))).... (-21.80 = -20.44 + -1.36)

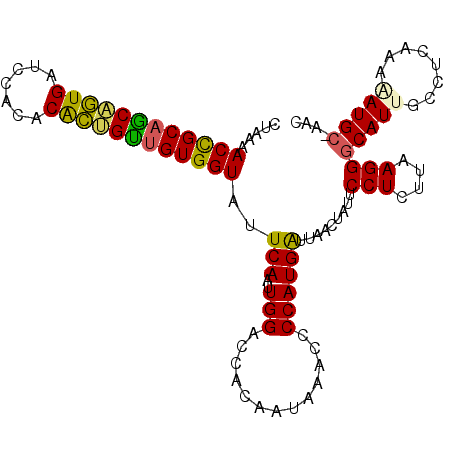

| Location | 16,979,557 – 16,979,651 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 70.53 |
| Mean single sequence MFE | -23.04 |
| Consensus MFE | -22.67 |
| Energy contribution | -21.11 |
| Covariance contribution | -1.56 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16979557 94 - 22224390 ---CACCAAGGGUAUCAUUAUCCGUCCAAAACUAAAACUGCAGCGUUGAUCCUUACAGGGUUGUGGUAUUCAACUGGACCAUAAUAAACCCCAUGAU ---......((((...(((((..(((((........((..((((.(((.......))).))))..)).......))))).)))))..))))...... ( -23.06) >DroVir_CAF1 6541 88 - 1 A-------ACACU--CGAUUUAUGUCCAAAACUAAAACCGCAUCAGUGAUUUACACGCUGGCGUGGUAUUCAAUUGGACUACAUAAAAGCCCAUGAU .-------.....--........((((((.......(((((..(((((.......)))))..)))))......)))))).................. ( -20.72) >DroGri_CAF1 5720 75 - 1 ----------------------UGUCCAAAACUAAAACCGCAUCAGUGAUUCACACGCUGAUGUGGUAUUCAAUUGGACUACAUCAAAGCCCAUGAU ----------------------.((((((.......((((((((((((.......))))))))))))......)))))).................. ( -25.72) >DroSim_CAF1 6492 94 - 1 ---CACCAAGAGUAUCAUUAUCCGUCCAAAACUAAAACUGCAGCGUUGAUCUCUACAGCGUUGUGGUAUUCAACUGGACCACAAUAAACCCCAUGAU ---....................(((((........((..((((((((.......))))))))..)).......))))).................. ( -20.86) >DroMoj_CAF1 5900 95 - 1 AUCUAUCAGAGAU--UGGCUUAUGUCCACAACUAAAACCGCAUCAGUGACCCACACGCUGGUGUGGUAUUCAAUUGGACUACAUUGAACCCCAUGGU ....((((.....--.((.(((.(((((........((((((((((((.......)))))))))))).......))))).....))).))...)))) ( -25.26) >DroAna_CAF1 9536 88 - 1 ---UGUAAA------CGAUAUCCGUCCAAAACUAAAACUGCAGCGUUGAUCCUCACAGCGUUGUGGUAUUCAAUUGGACCAAAAUGAACCCCAUGGU ---......------......((((((((.......((..((((((((.......))))))))..))......))))))....(((.....))))). ( -22.62) >consensus ____A_CAA___U__CGAUAUCCGUCCAAAACUAAAACCGCAGCAGUGAUCCACACACUGUUGUGGUAUUCAAUUGGACCACAAUAAACCCCAUGAU .......................(((((........((((((((((((.......)))))))))))).......))))).................. (-22.67 = -21.11 + -1.56)

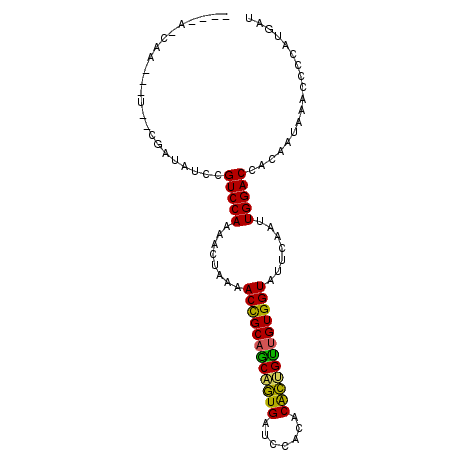

| Location | 16,979,651 – 16,979,756 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.68 |
| Mean single sequence MFE | -18.45 |
| Consensus MFE | -12.90 |
| Energy contribution | -13.07 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16979651 105 - 22224390 UGGACUAAUAUCAAUUGAAAUUGUUUUAACUAUCCUUGUGCUUGAAGAUAUAA---GCCACUUCCUGCUCUCCUGGAAAUUAAAACAACACUAAAGU-----UAAGCAUUAUU- ..((((....((....))..(((((((((...(((..(.((..((((......---....))))..)).)....)))..)))))))))......)))-----)..........- ( -15.30) >DroVir_CAF1 6629 104 - 1 UGGACUCAUAAUAAAU-AUAGGGUUUUAA-UAUCCUAGUGCCUGAAGAUCUCA---UUCUCUUCCCGCUCUCUUGGAACUUAAAACCACAUUACGCU----AUGCUCUUUUUG- .(((..((((......-....((((((((-..(((.((.((..(((((.....---...)))))..))...)).)))..)))))))).........)----))).))).....- ( -19.61) >DroPse_CAF1 6341 100 - 1 UGGACU-----CAAAGGAAACGGUUUUAA-UAUCCUAGUGCUUGAAGAACCAUAUCCCCUCUUCCCGCUCUCUUGGAACUUAAAACAACAUUU--------GUAAGCCCCAUCA .((...-----(((((....).(((((((-..(((.((.((..(((((...........)))))..))...)).)))..)))))))....)))--------)....))...... ( -19.60) >DroSim_CAF1 6586 105 - 1 UGGACUAAUAUCAAUUGAAAUUGUUUUAACUAUCCUUGUGCCUGAAGAUAUAA---GCCACUUCCUGCUCUCCUGGAAAUUAAAACAACACUAUAGU-----UAAGCAUUAUU- ..(((((...((....))..(((((((((...(((..(.((..((((......---....))))..)).)....)))..))))))))).....))))-----)..........- ( -15.80) >DroMoj_CAF1 5995 108 - 1 UGGACUGAUAUUAAAU-AUGAGGUUUUAA-UAUCCUAGUGCUUGAAGAUUAGA---CUCUCUUCCCGCUCUCUUGGAACUUAAAAUCACAUUUAUCUGAAUGUAAUCUUCAUU- ..((..(((.......-....((((((((-..(((.((.((..(((((.....---...)))))..))...)).)))..))))))))((((((....)))))).))).))...- ( -20.90) >DroPer_CAF1 6349 100 - 1 UGGACU-----CAAAGGAAACGGUUUUAA-UAUCCUAGUGCUUGAAGAUACAUAUCCCCUCUUCCCGCUCUCUUGGAACUUAAGACAACAUUU--------GUAAGCCCCACCA .((...-----(((((....).(((((((-..(((.((.((..(((((...........)))))..))...)).)))..)))))))....)))--------)....))...... ( -19.50) >consensus UGGACU_AUAUCAAAUGAAACGGUUUUAA_UAUCCUAGUGCUUGAAGAUAUAA___CCCUCUUCCCGCUCUCUUGGAACUUAAAACAACAUUA___U____GUAAGCAUCAUU_ .....................((((((((...(((.((.((..((((.............))))..))...)).)))..))))))))........................... (-12.90 = -13.07 + 0.17)


| Location | 16,979,721 – 16,979,821 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 69.32 |
| Mean single sequence MFE | -29.99 |
| Consensus MFE | -19.12 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.42 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16979721 100 + 22224390 GGAUAGUUAAAACAAUUUCAAUUGAUAUUAGUCCAUUUUUGGU-UAUGCUCAGUAUGGCAUCGUAUUUUUAA--GCCAUCUGGCAUUGAGGUGGACUGACACG ............((((....))))...((((((((((((....-.((((.(((.(((((.............--)))))))))))).)))))))))))).... ( -28.42) >DroVir_CAF1 6700 96 + 1 GGAUA-UUAAAACCCUAU-AUUUAUUAUGAGUCCAAUUGAAAUGUAUGCUUAGCUCAGCAUGGAAA-A-CAA--GCUGGCUGGCAUUGAGGUGGACUAAGAU- .....-............-..........((((((..........(((((.(((.((((.((....-.-)).--)))))))))))).....)))))).....- ( -23.96) >DroPse_CAF1 6412 94 + 1 GGAUA-UUAAAACCGUUUCCUUUG-----AGUCCAUUUUUAGUGGAUGCGCAGCU-GGGAAGGGGUUUUCCU--UUCAGCUGGCAUUGAGGUGGACUGACAAA (((..-...........)))....-----((((((((((.....(((((.(((((-((.(((((....))))--)))))))))))))))))))))))...... ( -33.12) >DroSim_CAF1 6656 99 + 1 GGAUAGUUAAAACAAUUUCAAUUGAUAUUAGUCCAUUUUU-UU-UAUGCUCAGUAUGGCAUGGUAUUUUUAA--GCCGUCUGGCAUUGAGGUGGACUGAAACG .....(((....((((....))))...((((((((((((.-..-.((((.(((.(((((.(((.....))).--)))))))))))).))))))))))))))). ( -31.10) >DroMoj_CAF1 6070 92 + 1 GGAUA-UUAAAACCUCAU-AUUUAAUAUCAGUCCAAUUGAGAU----GCGCAGUGCAG--UGGAAA-A-UAAUACUUGCCUGGCAUUGAGGUGGACUAACUU- .((((-(((((.......-.)))))))))((((((.....(((----((.(((.((((--((....-.-...))).)))))))))))....)))))).....- ( -30.20) >DroPer_CAF1 6420 94 + 1 GGAUA-UUAAAACCGUUUCCUUUG-----AGUCCAUUUUUAGUGGAUGCGCAGCU-GGGAAGGGGUUUUCCU--UUCAGCUGGCAUUGAGGUGGACUGACAAA (((..-...........)))....-----((((((((((.....(((((.(((((-((.(((((....))))--)))))))))))))))))))))))...... ( -33.12) >consensus GGAUA_UUAAAACCAUUUCAUUUGAUAU_AGUCCAUUUUUAGU_UAUGCGCAGCU_GGCAUGGAAUUUUCAA__GCCAGCUGGCAUUGAGGUGGACUGACAA_ .............................((((((((((......((((.(((.(((((....((....))...)))))))))))).))))))))))...... (-19.12 = -18.60 + -0.52)


| Location | 16,979,721 – 16,979,821 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 69.32 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -8.16 |
| Energy contribution | -8.66 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.37 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16979721 100 - 22224390 CGUGUCAGUCCACCUCAAUGCCAGAUGGC--UUAAAAAUACGAUGCCAUACUGAGCAUA-ACCAAAAAUGGACUAAUAUCAAUUGAAAUUGUUUUAACUAUCC .((((.((((((.....((((((((((((--.............))))).))).)))).-........)))))).))))........................ ( -21.86) >DroVir_CAF1 6700 96 - 1 -AUCUUAGUCCACCUCAAUGCCAGCCAGC--UUG-U-UUUCCAUGCUGAGCUAAGCAUACAUUUCAAUUGGACUCAUAAUAAAU-AUAGGGUUUUAA-UAUCC -.....((((((.....((((.(((((((--.((-.-....)).)))).)))..))))..........))))))..........-...((((.....-.)))) ( -19.36) >DroPse_CAF1 6412 94 - 1 UUUGUCAGUCCACCUCAAUGCCAGCUGAA--AGGAAAACCCCUUCCC-AGCUGCGCAUCCACUAAAAAUGGACU-----CAAAGGAAACGGUUUUAA-UAUCC ((((..((((((.....((((((((((.(--(((......))))..)-))))).))))..........))))))-----))))(....)........-..... ( -25.66) >DroSim_CAF1 6656 99 - 1 CGUUUCAGUCCACCUCAAUGCCAGACGGC--UUAAAAAUACCAUGCCAUACUGAGCAUA-AA-AAAAAUGGACUAAUAUCAAUUGAAAUUGUUUUAACUAUCC ......((((((.....(((((((..(((--.............)))...))).)))).-..-.....))))))............................. ( -16.84) >DroMoj_CAF1 6070 92 - 1 -AAGUUAGUCCACCUCAAUGCCAGGCAAGUAUUA-U-UUUCCA--CUGCACUGCGC----AUCUCAAUUGGACUGAUAUUAAAU-AUGAGGUUUUAA-UAUCC -.....((((((.....((((((((((.((....-.-.....)--)))).))).))----))......))))))(((((((((.-.......)))))-)))). ( -23.80) >DroPer_CAF1 6420 94 - 1 UUUGUCAGUCCACCUCAAUGCCAGCUGAA--AGGAAAACCCCUUCCC-AGCUGCGCAUCCACUAAAAAUGGACU-----CAAAGGAAACGGUUUUAA-UAUCC ((((..((((((.....((((((((((.(--(((......))))..)-))))).))))..........))))))-----))))(....)........-..... ( -25.66) >consensus _AUGUCAGUCCACCUCAAUGCCAGCCGAC__UUAAAAAUACCAUGCC_AACUGAGCAUA_ACUAAAAAUGGACU_AUAUCAAAUGAAACGGUUUUAA_UAUCC ......((((((.....(((((((..........................))).))))..........))))))............................. ( -8.16 = -8.66 + 0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:52 2006