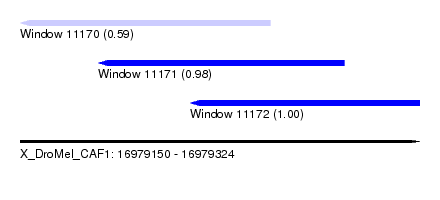
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,979,150 – 16,979,324 |
| Length | 174 |
| Max. P | 0.996995 |

| Location | 16,979,150 – 16,979,259 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.89 |
| Mean single sequence MFE | -20.38 |
| Consensus MFE | -16.95 |
| Energy contribution | -15.98 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
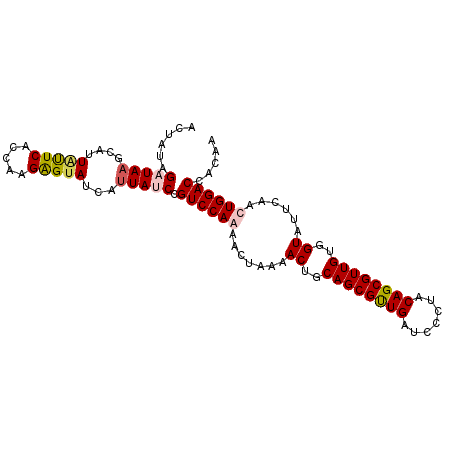
>X_DroMel_CAF1 16979150 109 - 22224390 ----UCCCUACAGCGUUGUGGUAUUC-AAUUGGACCACAAUAAACCCCAUGAUUAGCUAUUCCUCUUAAGGGCAUGGCCUCAAAAAUGCAACCUUAAUAAAGC-AAUAAAUCAAC----- ----..........((((((((....-......))))))))........(((((.(((((((((....)))).)))))........(((............))-)...)))))..----- ( -20.30) >DroSec_CAF1 7282 114 - 1 ----UCUCUACAGCGUUGUGGUAUUC-AAUUGGACCACAGGAAACCCCAUGGUUAGCUAUUCCUCUUAAGGGCAUGGCCUCAAAAAUGCAACCUUAAUAAAGC-AAUAAAUCAACAAAAC ----..........(((((..((((.-..((((((((..((....))..))))).(((((((((....)))).)))))..........)))....))))..))-)))............. ( -20.60) >DroSim_CAF1 6081 114 - 1 ----UCUCUACAGCGUUGUGGUAUUC-AACUGGACCACAAUAAACCCCAUGAUUAGCUAUUCCUCUUAAGGGCAUAGCCUCAAAAAUGAAACCUUAAUAAAGC-AAUAAAUCAACAAAAC ----........((((((((((....-......))))))))........(((...(((((((((....)))).))))).)))...................))-................ ( -20.00) >DroEre_CAF1 7373 114 - 1 ----UCUCUACAGCGUUGUGGUAUUC-AAUUGGACUACAACAAACCCCAUGAUUAGCUAUUCCUCUUAAGGGCAUGGGCUCAAAAAUGCAACCUUAAUAAAGC-AAUAAAUCAACAACAC ----........((((((((((....-......))))))))....((((((.........((((....)))))))))).......................))-................ ( -20.20) >DroWil_CAF1 85703 116 - 1 UAAAUCAAUAAAUUGUUGUAGUAUUCUAAAUGGACUACAACAAAC----UGAUAAACUACUCCUCUUAAGGAUAUCGUCUCGAAGAUAUAACCUUAAUAAAGUUCAAAAAUCAACAAAGU ............((((((((((..((.....))))))))))))..----((((.....(((....(((((((((((........)))))..))))))...)))......))))....... ( -20.80) >DroYak_CAF1 7923 114 - 1 ----UUCCCACAGCGUUGUGGUAUUC-AACUGGACUACAACAAACCCCAUGAUUAGCUAUUCCUCUUAAGGGCAUGGGCUCAAAAGUGCAACCUUAAUAAAGC-AAUAAAUCAACAAAAC ----..........((((((((....-......))))))))........(((((...((((.((.((((((...((.(((....))).)).))))))...)).-)))))))))....... ( -20.40) >consensus ____UCUCUACAGCGUUGUGGUAUUC_AAUUGGACCACAACAAACCCCAUGAUUAGCUAUUCCUCUUAAGGGCAUGGCCUCAAAAAUGCAACCUUAAUAAAGC_AAUAAAUCAACAAAAC ..............((((((((...........))))))))........(((((...........((((((((((..........))))..))))))...........)))))....... (-16.95 = -15.98 + -0.97)

| Location | 16,979,184 – 16,979,291 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -25.71 |
| Energy contribution | -25.22 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.37 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16979184 107 - 22224390 UUAUCCGUCCAAAACUAAAACUGCAGCGUUGA------------UCCCUACAGCGUUGUGGUAUUC-AAUUGGACCACAAUAAACCCCAUGAUUAGCUAUUCCUCUUAAGGGCAUGGCCU ......((((((.......((..((((((((.------------......))))))))..))....-..))))))....................(((((((((....)))).))))).. ( -29.52) >DroSec_CAF1 7321 103 - 1 ----CCGUCCAAAACUAAAACUGCAGCGUUGA------------UCUCUACAGCGUUGUGGUAUUC-AAUUGGACCACAGGAAACCCCAUGGUUAGCUAUUCCUCUUAAGGGCAUGGCCU ----((((((((.......((..((((((((.------------......))))))))..))....-..))))))....((.....))..))...(((((((((....)))).))))).. ( -31.62) >DroSim_CAF1 6120 107 - 1 UUAUCCGUCCAAAACUAAAACUGCAGCGUUGA------------UCUCUACAGCGUUGUGGUAUUC-AACUGGACCACAAUAAACCCCAUGAUUAGCUAUUCCUCUUAAGGGCAUAGCCU ......(((((........((..((((((((.------------......))))))))..))....-...)))))....................(((((((((....)))).))))).. ( -28.76) >DroEre_CAF1 7412 107 - 1 AUAUCCGUCCAAAACUAAAACUGCAGCGUUGA------------UCUCUACAGCGUUGUGGUAUUC-AAUUGGACUACAACAAACCCCAUGAUUAGCUAUUCCUCUUAAGGGCAUGGGCU ......((((((.......((..((((((((.------------......))))))))..))....-..))))))..........((((((.........((((....)))))))))).. ( -31.12) >DroWil_CAF1 85743 114 - 1 UU--ACGUCCAUCACUAAAACUAUAGCAAUGAGAUUUUCAUAAAUCAAUAAAUUGUUGUAGUAUUCUAAAUGGACUACAACAAAC----UGAUAAACUACUCCUCUUAAGGAUAUCGUCU ..--..((((((.......(((((((((((..(((((....))))).....))))))))))).......))))))..........----.((((......((((....)))))))).... ( -24.24) >DroYak_CAF1 7962 107 - 1 UUAUCCGUCCAAAACUAAAACUGCAGCGCUGA------------UUCCCACAGCGUUGUGGUAUUC-AACUGGACUACAACAAACCCCAUGAUUAGCUAUUCCUCUUAAGGGCAUGGGCU ......(((((........((..((((((((.------------......))))))))..))....-...)))))..........((((((.........((((....)))))))))).. ( -32.46) >consensus UUAUCCGUCCAAAACUAAAACUGCAGCGUUGA____________UCUCUACAGCGUUGUGGUAUUC_AAUUGGACCACAACAAACCCCAUGAUUAGCUAUUCCUCUUAAGGGCAUGGCCU ......((((((.......((((((((((((...................)))))))))))).......))))))....................(((((((((....)))).))))).. (-25.71 = -25.22 + -0.49)


| Location | 16,979,224 – 16,979,324 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -23.76 |
| Energy contribution | -24.57 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16979224 100 - 22224390 ACUAAAGUUAAGCAUUAUUCACCAAGGGUAUCAUUAUCCGUCCAAAACUAAAACUGCAGCGUUGAUCCCUACAGCGUUGUGGUAUUCAAUUGGACCACAA .........................(((((....)))))((((((.......((..((((((((.......))))))))..))......))))))..... ( -26.82) >DroSim_CAF1 6160 100 - 1 ACUAUAGUUAAGCAUUAUUCACCAAGAGUAUCAUUAUCCGUCCAAAACUAAAACUGCAGCGUUGAUCUCUACAGCGUUGUGGUAUUCAACUGGACCACAA ...............(((((.....))))).........(((((........((..((((((((.......))))))))..)).......)))))..... ( -22.76) >DroEre_CAF1 7452 100 - 1 ACUAUAGAUACACAUUACUCACCAAGAGGAUCAAUAUCCGUCCAAAACUAAAACUGCAGCGUUGAUCUCUACAGCGUUGUGGUAUUCAAUUGGACUACAA ......((((.......(((.....)))......)))).((((((.......((..((((((((.......))))))))..))......))))))..... ( -25.64) >DroYak_CAF1 8002 100 - 1 ACUAUAGAUAAGCAUUGUUCACCAAGAGUAUCAUUAUCCGUCCAAAACUAAAACUGCAGCGCUGAUUCCCACAGCGUUGUGGUAUUCAACUGGACUACAA ......((((..(.(((.....))))..)))).......(((((........((..((((((((.......))))))))..)).......)))))..... ( -27.56) >consensus ACUAUAGAUAAGCAUUAUUCACCAAGAGUAUCAUUAUCCGUCCAAAACUAAAACUGCAGCGUUGAUCCCUACAGCGUUGUGGUAUUCAACUGGACCACAA ......(((((....(((((.....)))))...))))).((((((.......((..((((((((.......))))))))..))......))))))..... (-23.76 = -24.57 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:48 2006