
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,978,044 – 16,978,134 |
| Length | 90 |
| Max. P | 0.665221 |

| Location | 16,978,044 – 16,978,134 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 74.11 |
| Mean single sequence MFE | -18.44 |
| Consensus MFE | -12.28 |
| Energy contribution | -12.00 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
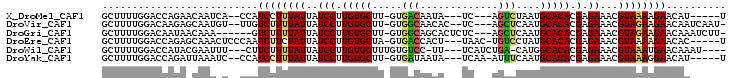
>X_DroMel_CAF1 16978044 90 - 22224390 GCUUUUGGACCAGAACAAUCA--CCAUCCUUUAUUAUCCUUGUGCUU-GUGACAAUA---UC---AGUCUAAUGCACACGAGAAACGUAAAAGAACAU-----U .((((((((...((....)).--...))).......(((.(((((..-..(((....---..---.)))....))))).).)).....))))).....-----. ( -13.00) >DroVir_CAF1 5272 95 - 1 GCUUUUGGACAAGAGCAAUGU--UUGUCUUUUAUUAUCCUUGUGCUU-GUGGCAACAC--UC---AGCUCAAUGCACACGAGAAACGUAGAAGAACAAUCAAU- ((((((....))))))..((.--(((((((((((....((((((...-(((....)))--..---.((.....)).))))))....))))))).)))).))..- ( -27.60) >DroGri_CAF1 4524 93 - 1 GCUUUUGGACAAUAACAAA------GUCUUUUAUUAUCCUUGUGCUU-GUGGCAGCACUCUC---AGCUCAAUGCACACGAGAAACGUAGAAGAACAAAUCUU- ..(((((........))))------)((((((((..(((.(((((((-(.(((.........---.)))))).))))).).))...)))))))).........- ( -18.40) >DroEre_CAF1 6259 94 - 1 GCUUUUGGACCAGAGCAAACUCCCAAUCUUCUAUUAUCCUUGUGCUA-GUGACCACU---UAAC-UGUCCUAUGCACACGAGAAACGUAAAAGAACAC-----U (((((......)))))..........((((.(((..(((.(((((((-(.(((....---....-.)))))).))))).).))...))).))))....-----. ( -17.90) >DroWil_CAF1 84375 92 - 1 GCUUUUGGACCAUACGAAUUU---CUUCUUUUAUUAUCCUUGUGCUUUGUGUCC-UU---UCAUCUGA-CAUGGCACACGAGAAACGUAAAUGAACAAAU---- .................((((---.(((.(((((....((((((..(..((((.-..---......))-))..)..))))))....))))).))).))))---- ( -15.80) >DroYak_CAF1 6664 92 - 1 GCUUUUGGACCAGAUUAAAUC--CCAUCCUUUAUUAUCCUUGUGCUU-GUGAUAAUA---UCAA-AUUUCAAUGCACACGAGAAACGUAAAGGAACAU-----U .....(((.............--)))((((((((..(((.(((((..-.(((.....---....-...)))..))))).).))...))))))))....-----. ( -17.92) >consensus GCUUUUGGACCAGAACAAACU__CCAUCUUUUAUUAUCCUUGUGCUU_GUGACAACA___UC___AGUUCAAUGCACACGAGAAACGUAAAAGAACAA______ ..........................((((((((..(((.(((((.....(((.............)))....))))).).))...)))))))).......... (-12.28 = -12.00 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:45 2006