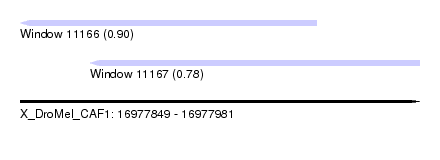
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,977,849 – 16,977,981 |
| Length | 132 |
| Max. P | 0.898783 |

| Location | 16,977,849 – 16,977,947 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -17.03 |
| Consensus MFE | -11.88 |
| Energy contribution | -10.97 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
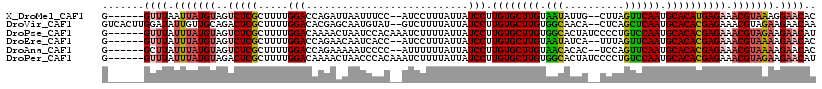
>X_DroMel_CAF1 16977849 98 - 22224390 UAAUUUCCAUCCUUUAUUAUCCUUGUGCUUGUAAUAUUGCUUAGUUCAAUGCACAUGAGAAACGUAAAGGAACACUAGCUUU-AAAUAAGUUAGGGACU---- .....(((.((((((((..(((.((((((((...((.....))...))).))))).).))...))))))))...(((((((.-....))))))))))..---- ( -26.50) >DroVir_CAF1 5082 96 - 1 CAAUGUAUGUCUUUUAUUAUCCUUGUGCUUGUGGCAACACUCAGCUCAAUGCACACGAGAAACGUAGAAGAACAAUU-------AAUAAGUCUAAGCAUGAAC .......((((((((((....((((((...(((....)))...((.....)).))))))....))))))).)))...-------................... ( -20.00) >DroSim_CAF1 4764 98 - 1 UAAUUCACAUCCUUUAUUAUCCUUGUGCUUGUAAUAUUACUUAGUUCAAUGCACACGAGAAACGUAAAUGAACACUAGCUUU-AAAUAAGUUAGGAACU---- ...(((...((.(((((..(((.((((((((...((.....))...))).))))).).))...))))).))...(((((((.-....))))))))))..---- ( -17.70) >DroEre_CAF1 6059 97 - 1 CAAUCACCAUCCUUUAUUAUCCUUGUGCUUGUAAUAUCAUUUAGUUCAAUGCACACGAGAAACGUAAAAGAACACUCGA-AU-AAAUAAAUUAGCAACU---- ............((((((.....((((((((...((.....))...))).)))))((((...............)))))-))-))).............---- ( -11.16) >DroYak_CAF1 6468 98 - 1 UAAUUCCCAUCCUUUAUUAUCCUUGUGCUUGCAAUAUCAUUCAGUUCAAUGCACACGAGAAACGUAAAAGAACACUAGCUUU-AAAUAAGUUGAGAACU---- .........((.(((((..(((.((((((((.(((........)))))).))))).).))...))))).))....((((((.-....))))))......---- ( -13.10) >DroAna_CAF1 8230 96 - 1 AAAUCCCCAUUUUUUAUUAUCCUUGUGCUUGUAACACACUCCAGUUCAAUGCACACGAGAAACGUAAAAGAACACUAUCUUCUAAAU---CUUAAAAAU---- .........((((((((..(((.((((((((.(((........)))))).))))).).))...))))))))................---.........---- ( -13.70) >consensus UAAUUCCCAUCCUUUAUUAUCCUUGUGCUUGUAAUAUCACUCAGUUCAAUGCACACGAGAAACGUAAAAGAACACUAGCUUU_AAAUAAGUUAAGAACU____ .........((.(((((..(((.((((((((.(((........)))))).))))).).))...))))).))................................ (-11.88 = -10.97 + -0.91)


| Location | 16,977,872 – 16,977,981 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.01 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -18.63 |
| Energy contribution | -18.66 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16977872 109 - 22224390 G------GUUUAAUUAUGUAGUCUCGCUUUUGGACCAGAUUAAUUUCC--AUCCUUUAUUAUCCUUGUGCUUGUAAUAUUG--CUUAGUUCAAUGCACAUGAGAAACGUAAAGGAACAC (------((((((....((......))..)))))))............--.((((((((..(((.((((((((...((...--..))...))).))))).).))...)))))))).... ( -23.00) >DroVir_CAF1 5103 115 - 1 GUCACUUGGAUAUUGUUGCAGACUCGCUUUUGGACACGAGCAAUGUAU--GUCUUUUAUUAUCCUUGUGCUUGUGGCAACA--CUCAGCUCAAUGCACACGAGAAACGUAGAAGAACAA .........((((((((((......))...((....)))))))))).(--(((((((((....((((((...(((....))--)...((.....)).))))))....))))))).))). ( -25.60) >DroPse_CAF1 4689 113 - 1 G------GUUUAUUUAUGUAGUCUCGCUUUUGGACAAAACUAAUCCACAAAUCUUUUAUUAUCCUUGUGCUUGUGGCACUAUCCCCUGUCCAAUGCACACGAGAAACGUAGAAGAACAU .------((((.(((((((..(((((....((((.........))))..................((((((((.((((........))))))).)))))))))).))))))).)))).. ( -28.30) >DroEre_CAF1 6081 109 - 1 G------GUUUAUUUAUGUAGUCUCGCUUUUGGACCAGAACAAUCACC--AUCCUUUAUUAUCCUUGUGCUUGUAAUAUCA--UUUAGUUCAAUGCACACGAGAAACGUAAAAGAACAC .------((((.(((((((..(((((.....(((...((....))...--.)))...........((((((((...((...--..))...))).)))))))))).))))))).)))).. ( -22.80) >DroAna_CAF1 8251 109 - 1 G------GCUUAUUUAUGUAGUCUCGCUUUUGGACCAGAAAAAUCCCC--AUUUUUUAUUAUCCUUGUGCUUGUAACACAC--UCCAGUUCAAUGCACACGAGAAACGUAAAAGAACAC .------.....(((((((..(((((.....(((...(((((((....--)))))))....))).((((((((.(((....--....)))))).)))))))))).)))))))....... ( -23.40) >DroPer_CAF1 4708 113 - 1 G------GUUUAUUUAUGUAGACUCGCUUUUGGACAAAACUAACCCACAAAUCUUUUAUUAUCCUUGUGCUUGUGGCACUAUCCCCUGUCCAAUGCACACGAGAAACGUAGAAGAACAU .------((((.(((((((...((((....(((...........)))..................((((((((.((((........))))))).)))))))))..))))))).)))).. ( -24.80) >consensus G______GUUUAUUUAUGUAGUCUCGCUUUUGGACAAGAAUAAUCCAC__AUCUUUUAUUAUCCUUGUGCUUGUAACACUA__CCCAGUUCAAUGCACACGAGAAACGUAAAAGAACAC .......((((.(((((((..(((((.....(((...........................))).((((((((.(((..........)))))).)))))))))).))))))).)))).. (-18.63 = -18.66 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:44 2006