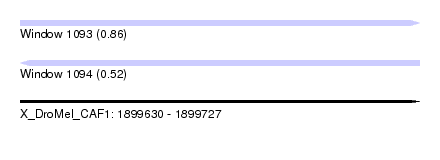
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,899,630 – 1,899,727 |
| Length | 97 |
| Max. P | 0.861883 |

| Location | 1,899,630 – 1,899,727 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -17.75 |
| Energy contribution | -17.87 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1899630 97 + 22224390 GGCACUGCCACUGAUUUAAA---UCACUUUUGUAUCAUUUUCAUUUCUGAUUGGUGCAUAAAUUAGAAUAU------UAUAUCAGUGGCACAUUUCAUUUUCGUUA .....((((((((((.(((.---...((..(((((((...(((....))).)))))))......))....)------)).))))))))))................ ( -23.50) >DroSec_CAF1 374 106 + 1 GGCACUACCACUGAUUUAAAUAAUCACUUUUGUAUCAUUUUCACUUCUGAUUGGUGCAGAAAUUCGAAUAUUCUCUUUAUAUCAGUGGCCCAUUUCGUUUUUGUUA ((.....((((((((.((((.......((((((((((...(((....))).))))))))))....((......)))))).)))))))).))............... ( -22.70) >DroSim_CAF1 398 106 + 1 GGCACUACUACUGAUUUAAAUAAUCACUUUUGUAUCAUUUUCACUUCUGAUUGGUGCAGAAAUUCGAAUAUUCUCUUUAUAUCAGUGGCCCAUUUCGUUUUCGUUA ((.....((((((((.((((.......((((((((((...(((....))).))))))))))....((......)))))).)))))))).))............... ( -20.40) >consensus GGCACUACCACUGAUUUAAAUAAUCACUUUUGUAUCAUUUUCACUUCUGAUUGGUGCAGAAAUUCGAAUAUUCUCUUUAUAUCAGUGGCCCAUUUCGUUUUCGUUA .......((((((((............((((((((((...(((....))).))))))))))......(((.......))))))))))).................. (-17.75 = -17.87 + 0.11)

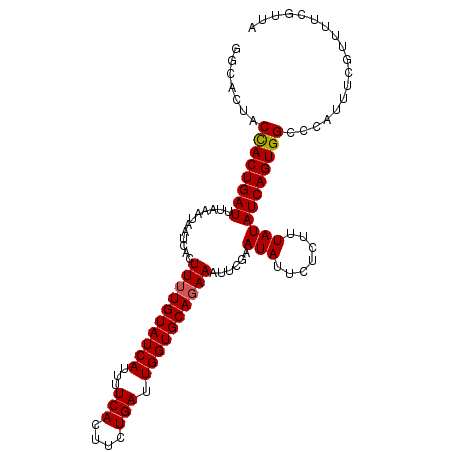
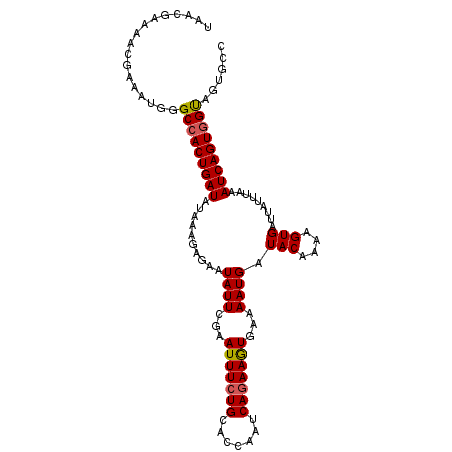
| Location | 1,899,630 – 1,899,727 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -18.77 |
| Consensus MFE | -16.54 |
| Energy contribution | -16.77 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1899630 97 - 22224390 UAACGAAAAUGAAAUGUGCCACUGAUAUA------AUAUUCUAAUUUAUGCACCAAUCAGAAAUGAAAAUGAUACAAAAGUGA---UUUAAAUCAGUGGCAGUGCC ................((((((((((...------.....................(((....)))((((.((......)).)---)))..))))))))))..... ( -20.10) >DroSec_CAF1 374 106 - 1 UAACAAAAACGAAAUGGGCCACUGAUAUAAAGAGAAUAUUCGAAUUUCUGCACCAAUCAGAAGUGAAAAUGAUACAAAAGUGAUUAUUUAAAUCAGUGGUAGUGCC .................(((((((((.................(((((((.......)))))))..(((((((((....)).)))))))..)))))))))...... ( -21.30) >DroSim_CAF1 398 106 - 1 UAACGAAAACGAAAUGGGCCACUGAUAUAAAGAGAAUAUUCGAAUUUCUGCACCAAUCAGAAGUGAAAAUGAUACAAAAGUGAUUAUUUAAAUCAGUAGUAGUGCC ................((((((((((.................(((((((.......)))))))..(((((((((....)).)))))))..))))))....).))) ( -14.90) >consensus UAACGAAAACGAAAUGGGCCACUGAUAUAAAGAGAAUAUUCGAAUUUCUGCACCAAUCAGAAGUGAAAAUGAUACAAAAGUGAUUAUUUAAAUCAGUGGUAGUGCC .................(((((((((..........((((...(((((((.......)))))))...)))).(((....))).........)))))))))...... (-16.54 = -16.77 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:59 2006