| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,930,802 – 16,931,002 |
| Length | 200 |
| Max. P | 0.854918 |

| Location | 16,930,802 – 16,930,922 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -42.52 |
| Consensus MFE | -31.82 |
| Energy contribution | -31.47 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16930802 120 + 22224390 CAUAACGUUGUACUAUGCCCUCAAGGACGGACUGCCGGAGGUUGAGGAACGCGCCCUCUGGACGAAUGGCUCCCAGCUGGCUCUCUUCUUCGGCACCGCCAUCUUUGCCUUCGAAGGCAU ..............(((((.((((((..(((.((.(((..(((((((((.(.(((..((((...........))))..))).)..))))))))).))).)))))...)))).)).))))) ( -39.70) >DroPse_CAF1 13700 120 + 1 CAUCACCCUCUACUAUGCUCUGAAGGAUGGACUGCCGGAGGUUAAGGAGCGCGCCUAUUGGACAAGCGGCUCCCAGUUGGCUCUCUUCUUUGGCACCGCCAUCUUUGCCUUUGAGGGCAU .....(((((......((....((((((((..((((((((((((((((((((.((....))....)).)))))...))))).)))).....))))...))))))))))....)))))... ( -42.50) >DroGri_CAF1 962 120 + 1 GAUAACAUUAUAUUAUGCACUGAAGGAUGGCUUGCCGGAGGUGAAGGAACGCGCCCUUUGGACGAAUGGUAGCCAGUUGGCGCUCUUCUUUGGCACCGCCAUCUUUGCCUUCGAAGGCAU ......................(((((((((.((((((((((......))(((((..((((.(........)))))..)))))....))))))))..)))))))))((((....)))).. ( -44.70) >DroWil_CAF1 520 120 + 1 CAUCACCUUGUACUAUGCCCUCAAAGAUGGACUGCCAGAGAUCGGGGAACGCGCCUACUGGACAAAUGGAUCGCAGUUGGCACUCUUCUUUGGCACCGCCAUAUUUGCCUUCGAGGGCAU ..............((((((((.(((((((..(((((((((..((((.....(((.((((((........)).)))).))).)))))))))))))...))))......))).)))))))) ( -42.60) >DroMoj_CAF1 591 120 + 1 CAUCACCCUCUACUACGCCCUCAAGGACGGCCUGCCGGAGGUGACGGAACGCGCCUACUGGACGAACGGCUCCCAGCUGGCGCUCUUCUUCGGCACCGCCAUCUUUGCCUUCGAGGGCAU ................((((((((((..(((.((((((((((......))(((((..((((...........))))..)))))....))))))))..))).......)))).)))))).. ( -49.60) >DroAna_CAF1 581 120 + 1 CAUAACCCUGUACUAUGCUUUGAAGGAUGGCCUGCCGGAUGUACCGGAUCGUGCUCUCUGGACCAAUGGCUCCCAGCUGGCCCUCUUCUUUGGCACGGCGAUCUUUGCUUAUGAGGGCAU .....(((((((....((....((((((.(((..((((.....))))...(((((....((.(((.(((...)))..))).))........)))))))).)))))))).))).))))... ( -36.00) >consensus CAUAACCCUGUACUAUGCCCUCAAGGAUGGACUGCCGGAGGUGAAGGAACGCGCCCACUGGACGAAUGGCUCCCAGCUGGCGCUCUUCUUUGGCACCGCCAUCUUUGCCUUCGAGGGCAU ......................((((((((..(((((((((....(((..(.(((..((((...........))))..))).)))))))))))))...))))))))((((....)))).. (-31.82 = -31.47 + -0.36)


| Location | 16,930,842 – 16,930,962 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -44.92 |
| Consensus MFE | -32.91 |
| Energy contribution | -33.05 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16930842 120 + 22224390 GUUGAGGAACGCGCCCUCUGGACGAAUGGCUCCCAGCUGGCUCUCUUCUUCGGCACCGCCAUCUUUGCCUUCGAAGGCAUUGCCUUAGUGAUGCCGUUGAAGAACGCCAUGCGAAAGCCA (((((((.......))))..)))...(((((....((((((..(((((..(((((.(((...(..(((((....)))))..).....))).)))))..)))))..)))).))...))))) ( -45.70) >DroPse_CAF1 13740 120 + 1 GUUAAGGAGCGCGCCUAUUGGACAAGCGGCUCCCAGUUGGCUCUCUUCUUUGGCACCGCCAUCUUUGCCUUUGAGGGCAUUGCACUGGUGAUGCCACUAAAGAAUGCGAUGCGAAAGCCA .....(((((((.((....))....)).)))))......((..((((...(((((.(((((.(..(((((....)))))..)...))))).)))))...))))..))...((....)).. ( -44.20) >DroGri_CAF1 1002 120 + 1 GUGAAGGAACGCGCCCUUUGGACGAAUGGUAGCCAGUUGGCGCUCUUCUUUGGCACCGCCAUCUUUGCCUUCGAAGGCAUUGCACUGGUGAUGCCCCUGAAGAAUGCCAUGCGCAAGUCG (((..((...(((((..((((.(........)))))..)))))(((((...((((.(((((.(..(((((....)))))..)...))))).))))...)))))...))...)))...... ( -45.60) >DroWil_CAF1 560 120 + 1 AUCGGGGAACGCGCCUACUGGACAAAUGGAUCGCAGUUGGCACUCUUCUUUGGCACCGCCAUAUUUGCCUUCGAGGGCAUUGCUCUGGUGAUGCCAUUGAAGAAUGCCAUGAGAAAACCC ...(((....(((((............))..)))...(((((.(((((..(((((.(((((....(((((....)))))......))))).)))))..))))).)))))........))) ( -45.70) >DroMoj_CAF1 631 120 + 1 GUGACGGAACGCGCCUACUGGACGAACGGCUCCCAGCUGGCGCUCUUCUUCGGCACCGCCAUCUUUGCCUUCGAGGGCAUUGCCCUGGUGAUGCCCCUGAAGAAUGCCAUGCGCAAGCCG ..((.(((..(((((..((((...........))))..)))))..))).))((((..((.......))(((((.((((((..(....)..)))))).)))))..))))..((....)).. ( -49.70) >DroAna_CAF1 621 120 + 1 GUACCGGAUCGUGCUCUCUGGACCAAUGGCUCCCAGCUGGCCCUCUUCUUUGGCACGGCGAUCUUUGCUUAUGAGGGCAUUGCCCUUGUGAUGCCCCUGAAGAACUCCAUGGCCAAGACC ((((......))))(((.(((.(((.(((......(((((((.........))).))))..((((((((((..(((((...)))))..))).))....)))))...)))))))))))).. ( -38.60) >consensus GUGAAGGAACGCGCCCACUGGACGAAUGGCUCCCAGCUGGCGCUCUUCUUUGGCACCGCCAUCUUUGCCUUCGAGGGCAUUGCCCUGGUGAUGCCCCUGAAGAAUGCCAUGCGAAAGCCA .....((..((((((..((((...........))))..)))..(((((...((((.(((((....(((((....)))))......))))).))))...))))).......)))....)). (-32.91 = -33.05 + 0.14)


| Location | 16,930,842 – 16,930,962 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -49.27 |
| Consensus MFE | -33.00 |
| Energy contribution | -34.92 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16930842 120 - 22224390 UGGCUUUCGCAUGGCGUUCUUCAACGGCAUCACUAAGGCAAUGCCUUCGAAGGCAAAGAUGGCGGUGCCGAAGAAGAGAGCCAGCUGGGAGCCAUUCGUCCAGAGGGCGCGUUCCUCAAC (((((..(((.((((.((((((..(((((((.(((...(..(((((....)))))..).))).)))))))..)))))).)))))).)..)))))........(((((.....)))))... ( -55.10) >DroPse_CAF1 13740 120 - 1 UGGCUUUCGCAUCGCAUUCUUUAGUGGCAUCACCAGUGCAAUGCCCUCAAAGGCAAAGAUGGCGGUGCCAAAGAAGAGAGCCAACUGGGAGCCGCUUGUCCAAUAGGCGCGCUCCUUAAC ((((((((((...)).(((((...(((((((.(((...(..((((......))))..).))).))))))))))))))))))))...(((((((((((((...))))))).)))))).... ( -53.30) >DroGri_CAF1 1002 120 - 1 CGACUUGCGCAUGGCAUUCUUCAGGGGCAUCACCAGUGCAAUGCCUUCGAAGGCAAAGAUGGCGGUGCCAAAGAAGAGCGCCAACUGGCUACCAUUCGUCCAAAGGGCGCGUUCCUUCAC .((...((((.((((.((((((...((((((.(((...(..(((((....)))))..).))).))))))...)))))).))))..............((((...))))))))....)).. ( -46.00) >DroWil_CAF1 560 120 - 1 GGGUUUUCUCAUGGCAUUCUUCAAUGGCAUCACCAGAGCAAUGCCCUCGAAGGCAAAUAUGGCGGUGCCAAAGAAGAGUGCCAACUGCGAUCCAUUUGUCCAGUAGGCGCGUUCCCCGAU (((........(((((((((((..(((((((.(((......((((......))))....))).)))))))..)))))))))))((((.(((......)))))))..........)))... ( -48.90) >DroMoj_CAF1 631 120 - 1 CGGCUUGCGCAUGGCAUUCUUCAGGGGCAUCACCAGGGCAAUGCCCUCGAAGGCAAAGAUGGCGGUGCCGAAGAAGAGCGCCAGCUGGGAGCCGUUCGUCCAGUAGGCGCGUUCCGUCAC (((..(((((.((((.((((((...((((((.(((...(..((((......))))..).))).))))))...)))))).))))(((((..(.....)..)))))..)))))..))).... ( -53.30) >DroAna_CAF1 621 120 - 1 GGUCUUGGCCAUGGAGUUCUUCAGGGGCAUCACAAGGGCAAUGCCCUCAUAAGCAAAGAUCGCCGUGCCAAAGAAGAGGGCCAGCUGGGAGCCAUUGGUCCAGAGAGCACGAUCCGGUAC (((....))).((((..(((((...(((((....(((((...))))).....((.......)).)))))...)))))(((((((.(((...))))))))))...........)))).... ( -39.00) >consensus CGGCUUGCGCAUGGCAUUCUUCAGGGGCAUCACCAGGGCAAUGCCCUCGAAGGCAAAGAUGGCGGUGCCAAAGAAGAGAGCCAACUGGGAGCCAUUCGUCCAGAAGGCGCGUUCCUUAAC .(((((.(...((((.((((((...((((((.(((......((((......))))....))).))))))...)))))).))))...).)))))...((((.....))))........... (-33.00 = -34.92 + 1.92)


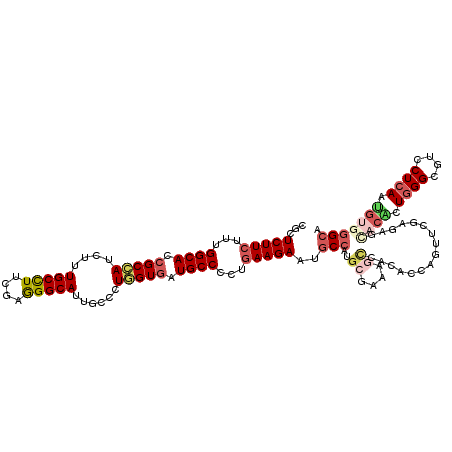
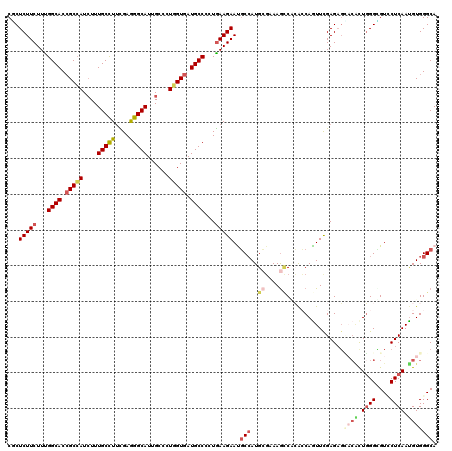
| Location | 16,930,882 – 16,931,002 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -45.50 |
| Consensus MFE | -29.68 |
| Energy contribution | -31.13 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16930882 120 + 22224390 CUCUCUUCUUCGGCACCGCCAUCUUUGCCUUCGAAGGCAUUGCCUUAGUGAUGCCGUUGAAGAACGCCAUGCGAAAGCCACACCAGUUCGAGCGACCCUUGGGUGUGCUCAAUGUGGGCA ...(((((..(((((.(((...(..(((((....)))))..).....))).)))))..)))))..(((..((....))..(((..(((.((((((((....))).)))))))))))))). ( -46.40) >DroPse_CAF1 13780 120 + 1 CUCUCUUCUUUGGCACCGCCAUCUUUGCCUUUGAGGGCAUUGCACUGGUGAUGCCACUAAAGAAUGCGAUGCGAAAGCCAAGCCAAUUCGAGAGCCGACUGGGGGUCCUCAAUGUGGGCA .((((.....(((((.(((((.(..(((((....)))))..)...))))).))))).....((((((...((....))...))..))))))))(((.((((((....))))..)).))). ( -45.90) >DroGri_CAF1 1042 120 + 1 CGCUCUUCUUUGGCACCGCCAUCUUUGCCUUCGAAGGCAUUGCACUGGUGAUGCCCCUGAAGAAUGCCAUGCGCAAGUCGCAACAAUUUGAGAGUACACUGGGCGUUCUCAAUGUGGGCA ...(((((...((((.(((((.(..(((((....)))))..)...))))).))))...))))).((((.((((.....)))).((..(((((((..(.....)..)))))))..)))))) ( -43.60) >DroWil_CAF1 600 120 + 1 CACUCUUCUUUGGCACCGCCAUAUUUGCCUUCGAGGGCAUUGCUCUGGUGAUGCCAUUGAAGAAUGCCAUGAGAAAACCCCACCAGUUCGAGAGCACAUUGGGUGUGCUCAAUGUGGGCA ...(((((..(((((.(((((....(((((....)))))......))))).)))))..))))).((((.((((...((((((...((.(....).))..)))).)).)))).....)))) ( -46.40) >DroMoj_CAF1 671 120 + 1 CGCUCUUCUUCGGCACCGCCAUCUUUGCCUUCGAGGGCAUUGCCCUGGUGAUGCCCCUGAAGAAUGCCAUGCGCAAGCCGCGCCAGUUUGAGAGCACGCUGGGCGUCCUCAAUGUGGGCA .((((......((((..((.......))(((((.((((((..(....)..)))))).)))))..))))..((....))(((.(((((.((....)).))))))))..........)))). ( -48.70) >DroAna_CAF1 661 120 + 1 CCCUCUUCUUUGGCACGGCGAUCUUUGCUUAUGAGGGCAUUGCCCUUGUGAUGCCCCUGAAGAACUCCAUGGCCAAGACCGAGCAGUUCGAGAUGACGUUCGGGGUCCUCAACGUGGGCA ........((((((..((.(.((((((((((..(((((...)))))..))).))....))))).).))...)))))).((((((.(((......))))))))).((((.......)))). ( -42.00) >consensus CGCUCUUCUUUGGCACCGCCAUCUUUGCCUUCGAGGGCAUUGCCCUGGUGAUGCCCCUGAAGAAUGCCAUGCGAAAGCCACACCAGUUCGAGAGCACACUGGGCGUCCUCAAUGUGGGCA ...(((((...((((.(((((....(((((....)))))......))))).))))...)))))..(((..((....))................((((.((((....)))).))))))). (-29.68 = -31.13 + 1.45)

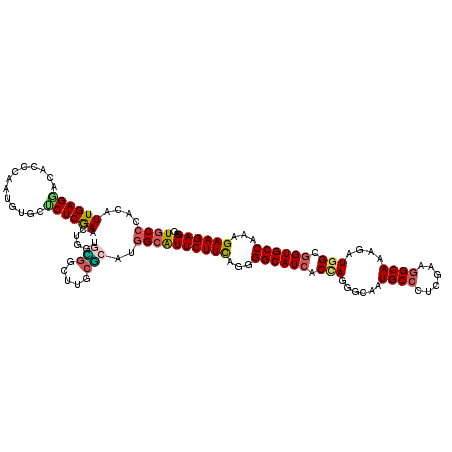
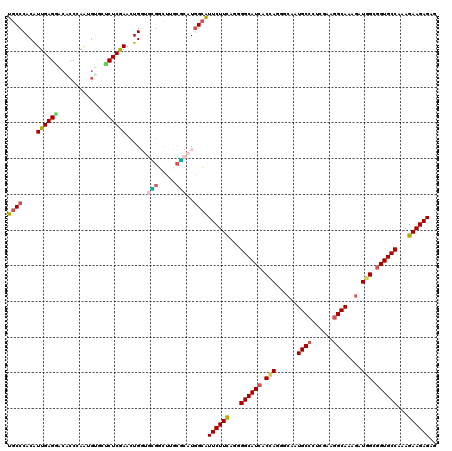
| Location | 16,930,882 – 16,931,002 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -43.83 |
| Consensus MFE | -29.10 |
| Energy contribution | -29.02 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16930882 120 - 22224390 UGCCCACAUUGAGCACACCCAAGGGUCGCUCGAACUGGUGUGGCUUUCGCAUGGCGUUCUUCAACGGCAUCACUAAGGCAAUGCCUUCGAAGGCAAAGAUGGCGGUGCCGAAGAAGAGAG .((((((.((((((..(((....))).))))))....))).)))...(((...)))((((((..(((((((.(((...(..(((((....)))))..).))).)))))))..)))))).. ( -47.30) >DroPse_CAF1 13780 120 - 1 UGCCCACAUUGAGGACCCCCAGUCGGCUCUCGAAUUGGCUUGGCUUUCGCAUCGCAUUCUUUAGUGGCAUCACCAGUGCAAUGCCCUCAAAGGCAAAGAUGGCGGUGCCAAAGAAGAGAG ((((((..(((((..((.......))..)))))..)))....((....))...)))((((((..(((((((.(((...(..((((......))))..).))).)))))))..)))))).. ( -40.20) >DroGri_CAF1 1042 120 - 1 UGCCCACAUUGAGAACGCCCAGUGUACUCUCAAAUUGUUGCGACUUGCGCAUGGCAUUCUUCAGGGGCAUCACCAGUGCAAUGCCUUCGAAGGCAAAGAUGGCGGUGCCAAAGAAGAGCG ((((....((((((((((...))))..)))))).....((((.....)))).))))((((((...((((((.(((...(..(((((....)))))..).))).))))))...)))))).. ( -43.80) >DroWil_CAF1 600 120 - 1 UGCCCACAUUGAGCACACCCAAUGUGCUCUCGAACUGGUGGGGUUUUCUCAUGGCAUUCUUCAAUGGCAUCACCAGAGCAAUGCCCUCGAAGGCAAAUAUGGCGGUGCCAAAGAAGAGUG ..(((((...(((((((.....)))))))........)))))............((((((((..(((((((.(((......((((......))))....))).)))))))..)))))))) ( -48.20) >DroMoj_CAF1 671 120 - 1 UGCCCACAUUGAGGACGCCCAGCGUGCUCUCAAACUGGCGCGGCUUGCGCAUGGCAUUCUUCAGGGGCAUCACCAGGGCAAUGCCCUCGAAGGCAAAGAUGGCGGUGCCGAAGAAGAGCG ((((......(((.((((...)))).)))........((((.....))))..))))((((((...((((((.(((...(..((((......))))..).))).))))))...)))))).. ( -45.90) >DroAna_CAF1 661 120 - 1 UGCCCACGUUGAGGACCCCGAACGUCAUCUCGAACUGCUCGGUCUUGGCCAUGGAGUUCUUCAGGGGCAUCACAAGGGCAAUGCCCUCAUAAGCAAAGAUCGCCGUGCCAAAGAAGAGGG (((((..((.((.(.((((((........))(((((.(..(((....)))..).)))))....))))).))))..)))))...(((((....(((..(....)..))).......))))) ( -37.60) >consensus UGCCCACAUUGAGGACACCCAAUGUGCUCUCGAACUGGUGCGGCUUGCGCAUGGCAUUCUUCAGGGGCAUCACCAGGGCAAUGCCCUCGAAGGCAAAGAUGGCGGUGCCAAAGAAGAGAG ((((....((((((.............))))))......(((.....)))..))))((((((...((((((.(((......((((......))))....))).))))))...)))))).. (-29.10 = -29.02 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:21 2006