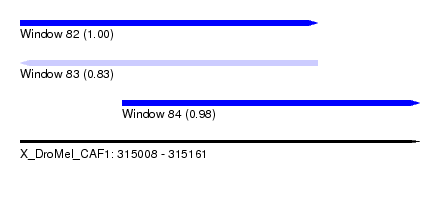
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 315,008 – 315,161 |
| Length | 153 |
| Max. P | 0.999768 |

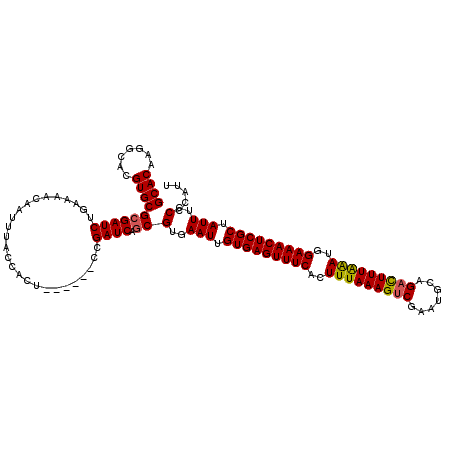
| Location | 315,008 – 315,122 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.64 |
| Mean single sequence MFE | -34.41 |
| Consensus MFE | -30.21 |
| Energy contribution | -30.13 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.04 |
| SVM RNA-class probability | 0.999768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 315008 114 + 22224390 GCACAAGGCACGUGCGCGAUCUGAAAACAAUUUACCACU------CCGAUCAGCGUGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGUUUUUAAAUAGAAACUCGCUAUUUCCCCAUG ((((((..(((((....((((.((..............)------).)))).)))))..))))(((((((..(((((((.(.......).)))))))..)))))))))............ ( -27.04) >DroSec_CAF1 57978 114 + 1 GCACAAGGCACGUGCGAGAUCUGAAAACAAUUUACCACU------CCGAUCAGCGUGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAAAUGGAAACUCGCUAUUUCCCCAUU ((((((..(((((....((((.((..............)------).)))).)))))..))))(((((((..(((((((((.......)))))))))..)))))))))............ ( -34.84) >DroSim_CAF1 78591 114 + 1 GCACAAGGCACGUGCGCGAUCUGAAAACAAUUUACCACU------CCGAUCAGCGUGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAAAUGGAAACUCGCUAUUUCCCCAUU ((((((..(((((....((((.((..............)------).)))).)))))..))))(((((((..(((((((((.......)))))))))..)))))))))............ ( -34.84) >DroEre_CAF1 58724 120 + 1 GCACAAGGCACGUGCGCGAUCUGAAAACAAUUUACCACUGCGAUCCCGAUCAGCGAGAAUUGUGAGUUUCACUUUAAAGUCGAACGCAGACUUUGAAGGGAAACUCGCCAUUUCCCCAUU ((.....(((.(((.(..(..((....))..)..)))))))(((....))).))(..(((.(((((((((.((((((((((.......)))))))))).))))))))).)))..)..... ( -37.30) >DroYak_CAF1 79482 114 + 1 GCACAAGGCACGUGCGCGAUCUGAAAACAAUUUACCACU------CCGAUCAGCGAGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAGAGGGAAACUCGCCAUUUCCCCAUU ((((.......))))((((((.((..............)------).)))).))(..(((.(((((((((.((((((((((.......)))))))))).))))))))).)))..)..... ( -38.04) >consensus GCACAAGGCACGUGCGCGAUCUGAAAACAAUUUACCACU______CCGAUCAGCGUGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAAAUGGAAACUCGCUAUUUCCCCAUU ((((.......))))((((((..........................)))).))(..(((.(((((((((..(((((((((.......)))))))))..))))))))).)))..)..... (-30.21 = -30.13 + -0.08)

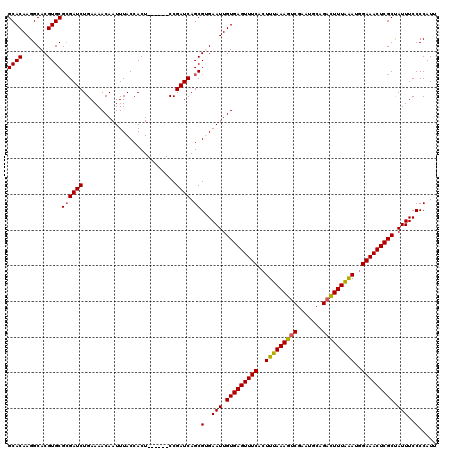
| Location | 315,008 – 315,122 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.64 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -27.72 |
| Energy contribution | -28.52 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 315008 114 - 22224390 CAUGGGGAAAUAGCGAGUUUCUAUUUAAAAACUGCAUUCGACUUUAAAGUGAAACUCACAAUUCACGCUGAUCGG------AGUGGUAAAUUGUUUUCAGAUCGCGCACGUGCCUUGUGC ((..(((.....(((((((((..((((((.............))))))..))))))).........((.((((.(------((............))).)))))))).....)))..)). ( -26.42) >DroSec_CAF1 57978 114 - 1 AAUGGGGAAAUAGCGAGUUUCCAUUUAAAGUCUGCAUUCGACUUUAAAGUGAAACUCACAAUUCACGCUGAUCGG------AGUGGUAAAUUGUUUUCAGAUCUCGCACGUGCCUUGUGC ....((((......(((((((..(((((((((.......)))))))))..)))))))((((((..((((......------))))...)))))).......))))(((((.....))))) ( -33.30) >DroSim_CAF1 78591 114 - 1 AAUGGGGAAAUAGCGAGUUUCCAUUUAAAGUCUGCAUUCGACUUUAAAGUGAAACUCACAAUUCACGCUGAUCGG------AGUGGUAAAUUGUUUUCAGAUCGCGCACGUGCCUUGUGC .(..(((.....(((((((((..(((((((((.......)))))))))..))))))).........((.((((.(------((............))).)))))))).....)))..).. ( -32.90) >DroEre_CAF1 58724 120 - 1 AAUGGGGAAAUGGCGAGUUUCCCUUCAAAGUCUGCGUUCGACUUUAAAGUGAAACUCACAAUUCUCGCUGAUCGGGAUCGCAGUGGUAAAUUGUUUUCAGAUCGCGCACGUGCCUUGUGC ....(((((.((..(((((((.(((.((((((.......)))))).))).))))))).)).)))))((.((((..((..(((((.....)))))..)).))))))(((((.....))))) ( -41.60) >DroYak_CAF1 79482 114 - 1 AAUGGGGAAAUGGCGAGUUUCCCUCUAAAGUCUGCAUUCGACUUUAAAGUGAAACUCACAAUUCUCGCUGAUCGG------AGUGGUAAAUUGUUUUCAGAUCGCGCACGUGCCUUGUGC ....(((((.((..(((((((.((.(((((((.......))))))).)).))))))).)).)))))((.((((.(------((............))).))))))(((((.....))))) ( -36.30) >consensus AAUGGGGAAAUAGCGAGUUUCCAUUUAAAGUCUGCAUUCGACUUUAAAGUGAAACUCACAAUUCACGCUGAUCGG______AGUGGUAAAUUGUUUUCAGAUCGCGCACGUGCCUUGUGC .(..(((.....(((((((((..(((((((((.......)))))))))..)))))))((((((..((((............))))...))))))...........)).....)))..).. (-27.72 = -28.52 + 0.80)

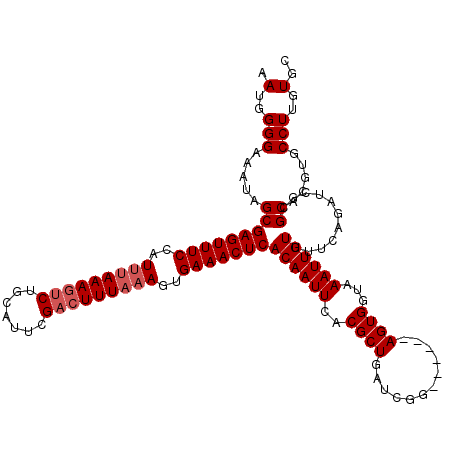
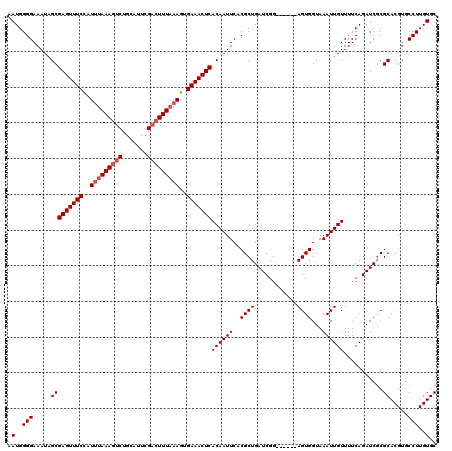
| Location | 315,047 – 315,161 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -23.10 |
| Energy contribution | -22.82 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 315047 114 + 22224390 -----CCGAUCAGCGUGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGUUUUUAAAUAGAAACUCGCUAUUUCCCCAUGU-AUUGUUUAAAAGAGGUAUUAAAACAGCCCGCACCCGCU -----......((((.(((..(((((((((..(((((((.(.......).)))))))..)))))))))...))).....(.-.((((((.............))))))..).....)))) ( -21.22) >DroSec_CAF1 58017 102 + 1 -----CCGAUCAGCGUGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAAAUGGAAACUCGCUAUUUCCCCAUUC-A---UUUCAAAGCGGCAUUACAACUG---------GCU -----......((((..(((.(((((((((..(((((((((.......)))))))))..))))))))).)))..)......-.---........(((........)))---------))) ( -26.80) >DroSim_CAF1 78630 102 + 1 -----CCGAUCAGCGUGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAAAUGGAAACUCGCUAUUUCCCCAUUC-A---UUUCAAAGCGGCAUUACAACUG---------GCU -----......((((..(((.(((((((((..(((((((((.......)))))))))..))))))))).)))..)......-.---........(((........)))---------))) ( -26.80) >DroEre_CAF1 58764 120 + 1 CGAUCCCGAUCAGCGAGAAUUGUGAGUUUCACUUUAAAGUCGAACGCAGACUUUGAAGGGAAACUCGCCAUUUCCCCAUUUCAUUGUUUUAAAACGGCAUUAGAACUGGUCGCAGCCACU .(((....))).((((((((.(((((((((.((((((((((.......)))))))))).))))))))).))))..(((.(((..((((.......))))...))).)))))))....... ( -38.60) >DroYak_CAF1 79521 91 + 1 -----CCGAUCAGCGAGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAGAGGGAAACUCGCCAUUUCCCCAUUUCAUUGUUUCAAAGCG------------------------ -----.......(((..(((.(((((((((.((((((((((.......)))))))))).))))))))).)))..).((......)).......)).------------------------ ( -28.80) >consensus _____CCGAUCAGCGUGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAAAUGGAAACUCGCUAUUUCCCCAUUU_AUUGUUUCAAAGCGGCAUUACAACUG_________GCU ............(.(..(((.(((((((((..(((((((((.......)))))))))..))))))))).)))..).)........................................... (-23.10 = -22.82 + -0.28)

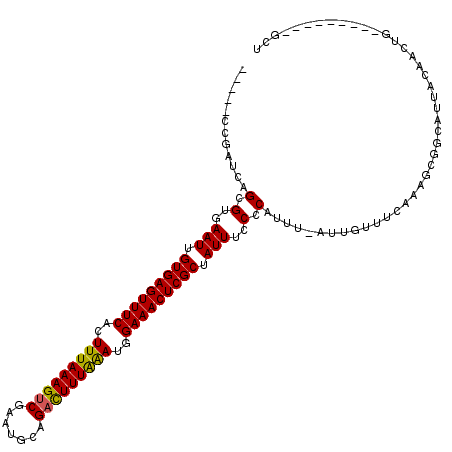
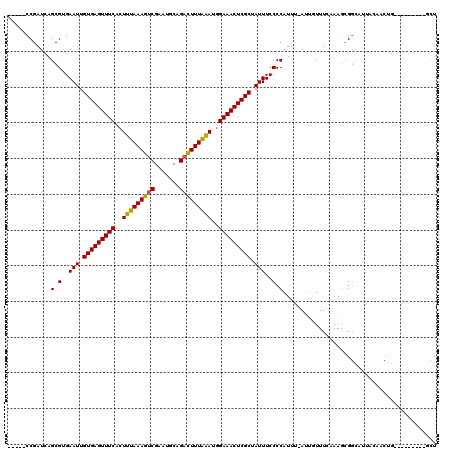
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:55 2006