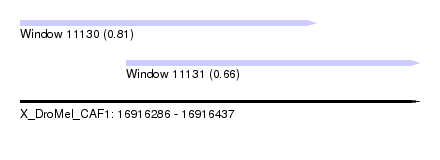
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,916,286 – 16,916,437 |
| Length | 151 |
| Max. P | 0.809778 |

| Location | 16,916,286 – 16,916,398 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.58 |
| Mean single sequence MFE | -48.97 |
| Consensus MFE | -31.85 |
| Energy contribution | -31.22 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
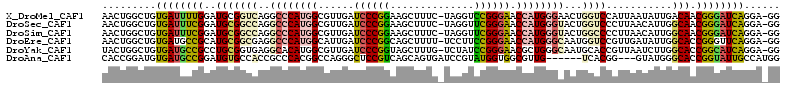
>X_DroMel_CAF1 16916286 112 + 22224390 GCAGUCUGGGCGUUGCAUGCGAUGAGGACUGUGGCUAUGGAACUGGCUGUGAUUUUGGAUGCGGUCAGGCCCAUGGCGUUGAUCCCGGAAGCUUUC-UAGGUCCGGGAACCAU (((((((...(((((....))))).))))))).(((((((..((((((((..........))))))))..))))))).....(((((((..((...-.)).)))))))..... ( -52.00) >DroSec_CAF1 3291 112 + 1 GCAGUCUGGGCGUUGCAUGCGAUGAGGACUGUGGCUAUGGAACUGGCUGUGAUUUCGGAUGCGGCCAGGCCCAUGGCGUUGAUCCCGGAAGCUUUC-UAGGUUCGGGAACCAU (((((((...(((((....))))).))))))).(((((((..((((((((..........))))))))..))))))).....(((((((..((...-.)).)))))))..... ( -51.80) >DroSim_CAF1 3238 112 + 1 GCAGUCUGGGCGUUGCAUGCGAUGAGGACUGUGGCUAUGGAACUGGCUGUGAUUUCGGAUGCGGCCAGGCCCAUGGCGUUGAUCCCGGAAGCUUUC-UAGGUUCGGGAACCAU (((((((...(((((....))))).))))))).(((((((..((((((((..........))))))))..))))))).....(((((((..((...-.)).)))))))..... ( -51.80) >DroEre_CAF1 3296 112 + 1 GCAGCCUGGGCAUUGCGUGCGAUGAGGACUGUGGCUAUGGAACUGGCUGUGAUGCCGCAUGCGGCGAGGCCCAUGGCAUUGAUCCCGGCAGCUUUU-UCCUUCCGGGAACCAU ...((((((((.((((.(((.(((.((..((..((((......))))..))...)).)))))))))).))))).))).....((((((.((.....-..)).))))))..... ( -45.40) >DroYak_CAF1 3272 112 + 1 GCAGCCUGGGCGUGGCAUGCGACGAGGACUGCGGCUAUGGUACUGGCUGUGAUGCCGCCUGCGGUGAGGCACAUGGCGUUGAUCCCGGUAGCUUUG-UCUAUCCGGGAACGCU (((((((.(.(((.....))).).))).)))).((((((.....(((......)))((((......)))).)))))).....(((((((((.....-.))).))))))..... ( -46.80) >DroAna_CAF1 3172 113 + 1 GCUCCCUGGGUGCGCCCUGUGACGAGGAUUGCGGCUACGGCACCGGAUGUGAUGCCGGAUGUGCCACCGCCCACGGCCAGGGCUCCGUCAGCAGUGAUCCGUAUGGUGGCGUU (((.....)))((((((((((....((((..(.(((((((((((((........))))...))))...((((.......))))...)).))).)..)))).))))).))))). ( -46.00) >consensus GCAGCCUGGGCGUUGCAUGCGAUGAGGACUGUGGCUAUGGAACUGGCUGUGAUGCCGGAUGCGGCCAGGCCCAUGGCGUUGAUCCCGGAAGCUUUC_UAGGUCCGGGAACCAU (((((((.(.(((.....))).).))).)))).(((((((..((.(((((..........))))).))..))))))).....((((((..............))))))..... (-31.85 = -31.22 + -0.63)



| Location | 16,916,326 – 16,916,437 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.97 |
| Mean single sequence MFE | -44.85 |
| Consensus MFE | -24.19 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16916326 111 + 22224390 AACUGGCUGUGAUUUUGGAUGCGGUCAGGCCCAUGGCGUUGAUCCCGGAAGCUUUC-UAGGUCCGGGAACCAUGGGAACUGGUCCAUUAAUAUUGACAACGGGAUCAGGA-GG ..((((((((..........)))))))).(((((((......(((((((..((...-.)).))))))).)))))))..(((((((................)))))))..-.. ( -43.09) >DroSec_CAF1 3331 111 + 1 AACUGGCUGUGAUUUCGGAUGCGGCCAGGCCCAUGGCGUUGAUCCCGGAAGCUUUC-UAGGUUCGGGAACCAUGGGUACUGGUCCCUUAACAUUGGCAACGGGAUCAGGA-GG ..((((((((..........))))))))((((((((......(((((((..((...-.)).))))))).)))))))).((((((((........(....)))))))))..-.. ( -48.20) >DroSim_CAF1 3278 111 + 1 AACUGGCUGUGAUUUCGGAUGCGGCCAGGCCCAUGGCGUUGAUCCCGGAAGCUUUC-UAGGUUCGGGAACCAUGGGUACUGGCCCCUUAACAUUGGCAACGGGAUCAGGA-GG .......(.((((..((..(((((((((((((((((......(((((((..((...-.)).))))))).)))))))).)))))).(........)))).))..)))).).-.. ( -46.30) >DroEre_CAF1 3336 111 + 1 AACUGGCUGUGAUGCCGCAUGCGGCGAGGCCCAUGGCAUUGAUCCCGGCAGCUUUU-UCCUUCCGGGAACCAUGGGCAAUGGUCCGUUGAUAUUGGCACCGGGUUCAGGA-GG ..((((((....(((((....)))))..((((((((......((((((.((.....-..)).)))))).))))))))..(((((((.......))).)))))).))))..-.. ( -43.20) >DroYak_CAF1 3312 111 + 1 UACUGGCUGUGAUGCCGCCUGCGGUGAGGCACAUGGCGUUGAUCCCGGUAGCUUUG-UCUAUCCGGGAACGCUGGGCAAUGCACCGUUAAUCUUGGCACCGGCAUCAGGA-GG .......(.((((((((((.((((((..((...((((((...(((((((((.....-.))).)))))))))))).))....)))))).......)))...))))))).).-.. ( -45.20) >DroAna_CAF1 3212 104 + 1 CACCGGAUGUGAUGCCGGAUGUGCCACCGCCCACGGCCAGGGCUCCGUCAGCAGUGAUCCGUAUGGUGGCGUUG------UCACGG---GUAUGGGCACCGGUAUUGCCAUGG ..(((...(..(((((((...((((...((((((((.((..(((.....)))..))..))))...(((((...)------))))))---))...)))))))))))..)..))) ( -43.10) >consensus AACUGGCUGUGAUGCCGGAUGCGGCCAGGCCCAUGGCGUUGAUCCCGGAAGCUUUC_UAGGUCCGGGAACCAUGGGCACUGGUCCGUUAAUAUUGGCAACGGGAUCAGGA_GG .........((((((((..(((((((..((((((((......((((((..............)))))).))))))))...))))...........))).))))))))...... (-24.19 = -24.92 + 0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:10 2006