
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,912,651 – 16,912,807 |
| Length | 156 |
| Max. P | 0.999270 |

| Location | 16,912,651 – 16,912,767 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.02 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -20.82 |
| Energy contribution | -22.50 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16912651 116 + 22224390 AAAAUAGUUGAAGAAAUUCUAGUGAUUAACCUAAAAACAUCGAUAGUCUGAAAUAAGCAUCGAUGUUUGGAAUAUUUAAAAAAAAACCUCGGUGCAUCGAUAUU----AGCAUCGAUGUU .........(((....)))................(((((((((.(.((((.((..(((((((.((((...............)))).)))))))....)).))----)))))))))))) ( -26.06) >DroSec_CAF1 7997 107 + 1 AAAAUAGU----------UAGGUGAUUAACCUAAAGAUAUCGAUAGUCUGAAAUAAACAUCGAUGUGCGGGAUAAAUAAA---AAACAUCGGUGCAUCGAUAUUAGUUAGCAUAGAUGUU .......(----------(((((.....)))))).((((((....(.((((.......(((((((..(.((.........---.....)).)..))))))).....)))))...)))))) ( -23.74) >DroSim_CAF1 8501 103 + 1 -----AUU----------UAGGUGAUUAACCUAAAGAUAUCGAUAGUCUGAAAUAAACAUCGAUGUGCGGCAUAAAUAAA--AAAACAUCGGUGCAUCGAUAUUAGUUAGCAUCGAUGUU -----.((----------(((((.....)))))))(((((((((.(.((((.......(((((((..(.(..........--.......).)..))))))).....)))))))))))))) ( -29.53) >consensus AAAAUAGU__________UAGGUGAUUAACCUAAAGAUAUCGAUAGUCUGAAAUAAACAUCGAUGUGCGGAAUAAAUAAA__AAAACAUCGGUGCAUCGAUAUUAGUUAGCAUCGAUGUU ..................(((((.....)))))..(((((((((.(.((((....((.((((((((((.((.................)).)))))))))).))..)))))))))))))) (-20.82 = -22.50 + 1.68)

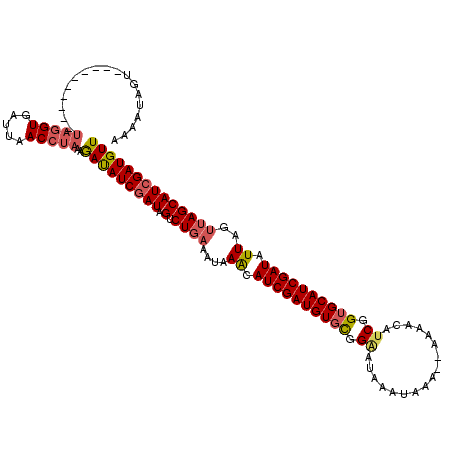

| Location | 16,912,651 – 16,912,767 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.02 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -15.05 |
| Energy contribution | -15.61 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16912651 116 - 22224390 AACAUCGAUGCU----AAUAUCGAUGCACCGAGGUUUUUUUUUAAAUAUUCCAAACAUCGAUGCUUAUUUCAGACUAUCGAUGUUUUUAGGUUAAUCACUAGAAUUUCUUCAACUAUUUU ..((((((((..----..))))))))....((((...((((.........(((((((((((((.((......)).)))))))))))...)).........))))...))))......... ( -24.87) >DroSec_CAF1 7997 107 - 1 AACAUCUAUGCUAACUAAUAUCGAUGCACCGAUGUUU---UUUAUUUAUCCCGCACAUCGAUGUUUAUUUCAGACUAUCGAUAUCUUUAGGUUAAUCACCUA----------ACUAUUUU ((((((..(((..............)))..)))))).---................((((((((((.....)))).))))))....((((((.....)))))----------)....... ( -18.64) >DroSim_CAF1 8501 103 - 1 AACAUCGAUGCUAACUAAUAUCGAUGCACCGAUGUUUU--UUUAUUUAUGCCGCACAUCGAUGUUUAUUUCAGACUAUCGAUAUCUUUAGGUUAAUCACCUA----------AAU----- (((((((.(((..............))).)))))))..--................((((((((((.....)))).))))))...(((((((.....)))))----------)).----- ( -22.64) >consensus AACAUCGAUGCUAACUAAUAUCGAUGCACCGAUGUUUU__UUUAUUUAUCCCGCACAUCGAUGUUUAUUUCAGACUAUCGAUAUCUUUAGGUUAAUCACCUA__________ACUAUUUU (((((((.(((..............))).)))))))....................(((((((.((......)).))))))).....(((((.....))))).................. (-15.05 = -15.61 + 0.56)

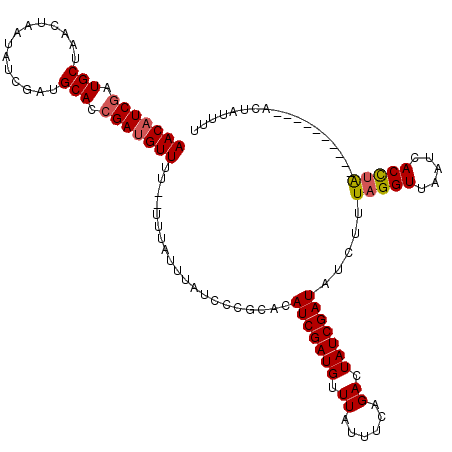
| Location | 16,912,691 – 16,912,807 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.78 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -24.54 |
| Energy contribution | -26.43 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16912691 116 - 22224390 UAUUAUCGGUAUCGGGAUGAGUUUUUAAUAGAGGUGCCGAAACAUCGAUGCU----AAUAUCGAUGCACCGAGGUUUUUUUUUAAAUAUUCCAAACAUCGAUGCUUAUUUCAGACUAUCG .......((((((((...((((.(((((.(((((.(((....((((((((..----..))))))))......))).)))))))))).))))......))))))))............... ( -25.10) >DroSec_CAF1 8027 117 - 1 UAUUAUCGGUAUCGGGAUGAGUUUUUAAUAGAGGUGCCAAAACAUCUAUGCUAACUAAUAUCGAUGCACCGAUGUUU---UUUAUUUAUCCCGCACAUCGAUGUUUAUUUCAGACUAUCG ...(((((((..((((((((((....((((..(((((......(((.(((........))).))))))))..)))).---...))))))))))...)))))))................. ( -30.20) >DroSim_CAF1 8526 118 - 1 UAUUAUCGGUAUCGGGAUGAGUUUUUAAUAGAGGUGCCAAAACAUCGAUGCUAACUAAUAUCGAUGCACCGAUGUUUU--UUUAUUUAUGCCGCACAUCGAUGUUUAUUUCAGACUAUCG ...(((((((..(((.((((((....((((..(((((......(((((((........))))))))))))..))))..--...)))))).)))...)))))))................. ( -30.80) >consensus UAUUAUCGGUAUCGGGAUGAGUUUUUAAUAGAGGUGCCAAAACAUCGAUGCUAACUAAUAUCGAUGCACCGAUGUUUU__UUUAUUUAUCCCGCACAUCGAUGUUUAUUUCAGACUAUCG ...(((((((..((((((((((....((((..(((((......(((((((........))))))))))))..)))).......))))))))))...)))))))................. (-24.54 = -26.43 + 1.89)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:05 2006