| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,906,383 – 16,906,490 |
| Length | 107 |
| Max. P | 0.801611 |

| Location | 16,906,383 – 16,906,490 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
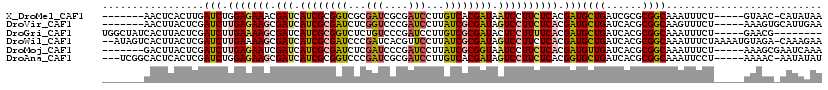
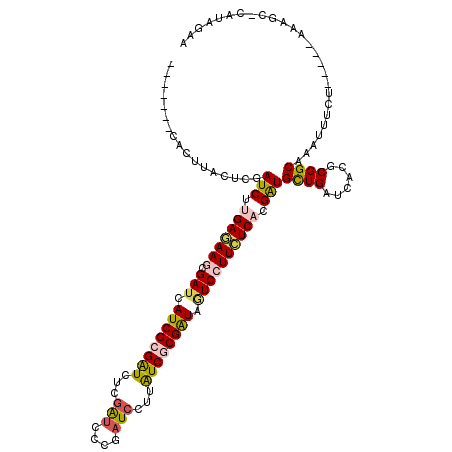
>X_DroMel_CAF1 16906383 107 + 22224390 -------AACUCACUUGAUCUGGAGAAACGAUCAUCGCGGUCGCGAUCGCGAUCCUUGUCACGAUAAUCCUUCUCACGAUGCUGAUCGCGCGGCAAAUUUCU-----GUAAC-CAUAUAA -------........(((((.(......))))))..((.(.((((((((((((....)))......(((........))))).)))))))).))........-----.....-....... ( -23.00) >DroVir_CAF1 2574 108 + 1 -------AACUUACUCGAUCUUGAGAAGCGAUCAUCGCGAUCUCGGUCCCGAUCCUUAUCGCGAUAGUCCUUCUCACGAUGCUGAUCACGCGGCAAGUUUCU-----AAAGUGCAUUGAA -------(((((.....(((.(((((((.(((.((((((((.(((....))).....)))))))).)))))))))).)))((((......)))))))))...-----............. ( -34.60) >DroGri_CAF1 3268 107 + 1 UGGCUAUCACUUACUCGAUCUUGAAAAGCGAUCAUCGCGGUCUCUGUCCCGAUCCUUGUCGCGAUACUCCUUUUCACGAUGCUGAUCACGCGGCAAAUUUCU-----GAACG-------- ......(((........(((.(((((((.((..(((((((........))(((....))))))))..))))))))).)))((((......)))).......)-----))...-------- ( -24.40) >DroWil_CAF1 2582 117 + 1 --AUAGUCACUUACUCGAUCUUGAAAAGCGAUCAUCGCGAUCCCGAUCACGUUCCUUAUCGCGAUAGUCCUUCUCACGAUGCUGAUCACGCGGCAAAUUUCUAAAAUGUAGA-CAAAGAA --...(((.........(((.(((.(((.(((.((((((((...((......))...)))))))).)))))).))).)))((((......))))................))-)...... ( -29.30) >DroMoj_CAF1 2401 108 + 1 -------GACUUACUCGAUCUUGAGAAUCGAUCAUCGCGAUCUCGAUCCCGAUCCUUAUCGCGGUAAUCCUUCUCACGAUGUUGAUCACGCGGCAAAUUUCU-----AAAGCGAAUCAAA -------..........(((.((((((..(((.((((((((...(((....)))...)))))))).))).)))))).))).(((((..(((...........-----...))).))))). ( -28.24) >DroAna_CAF1 2195 111 + 1 ---UCGGCACUCACUCGAUCUGGAGAAGCGAUCAUCGCGGUCCCGAUCGCGAUCCUUGUCACGAUAGUCCUUCUCACGGUGCUGAUCACGCGGCAAAUUCCU-----AAAAC-AAUAUAU ---((((((((..(.......)((((((.(((.((((((((....)))))(((....)))..))).)))))))))..)))))))).................-----.....-....... ( -29.60) >consensus _______CACUUACUCGAUCUUGAGAAGCGAUCAUCGCGAUCUCGAUCCCGAUCCUUAUCGCGAUAGUCCUUCUCACGAUGCUGAUCACGCGGCAAAUUUCU_____AAAGC_CAUAGAA .................(((.(((((((.(((.((((((((...(((....)))...)))))))).)))))))))).)))((((......)))).......................... (-23.34 = -23.48 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:01 2006