
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,891,150 – 16,891,299 |
| Length | 149 |
| Max. P | 0.598189 |

| Location | 16,891,150 – 16,891,265 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.68 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -28.64 |
| Energy contribution | -28.92 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16891150 115 - 22224390 CUUAACCGGCUGAGCUUAAUUGGUUUGCCUUGCCAACGAGCUGGGCGGCAUUUUCCGAUUAAUUGAUGGGUGGGAAAAGUGUGU-----GGACGAGACGAAAGUCAAGUUGCCAUUUAAA ........(((.(((((..(((((.......))))).))))).)))(((.((((((.(((........))).))))))......-----.(((..(((....)))..))))))....... ( -37.10) >DroSec_CAF1 90121 115 - 1 CUUAACCGGCUGAGCUUAAUUGGUUUGCCUUGCCAACGAGCUGGGCGGCAUUUUCCGAUUAAUUGAUGGGUGGGAAAAGUGUGU-----GGACGAGACGAAAGUCAAGUCGGCAUUUAAA .....(((.((.(((((..(((((.......))))).))))).)))))..((((((.(((........))).))))))..((((-----.(((..(((....)))..))).))))..... ( -39.70) >DroSim_CAF1 70568 115 - 1 CUUAACCGGCUGAGCUUAAUUGGUUUGCCUUGCCAACGAGCUGGGCGGUAUUUUCCGAUUAAUUGAUGGGUGGGAAAAGUGUGU-----GGACGAGACGAAAGUCAAGUCGGCAUUUAAA ....((((.((.(((((..(((((.......))))).))))).)))))).((((((.(((........))).))))))..((((-----.(((..(((....)))..))).))))..... ( -41.50) >DroEre_CAF1 74614 114 - 1 CUUAACCGGCUGAGCUUAAUUGGUUUGCCUUGCCGACGAGCUGGGCGGCAUUUUCCGAUUAAUUGAUGGGUGGGAAAAGUGUGC-----GGACGCAAGGAAAGUCAA-CUGCCAUUUAAA ........(((.(((((..(((((.......))))).))))).)))((((((((((.(((........))).))))))((((..-----..))))............-.))))....... ( -32.50) >DroYak_CAF1 76588 120 - 1 CUUAACCGGCUGAGCUUAAUUGGUUUGCCUUGCCAACGAGCUGGGCGGCAUUUUCCGAUUAACUGAUGGGUGGGAAAAGUGUGCGAUACGGACGAAACGAAAGUCAAGCUGGCAUUUAAA .....((((((..((((..((.((((((((.((......)).)))).(((((((((.(((........))).)))))...)))).....)))).))....))))..))))))........ ( -33.00) >consensus CUUAACCGGCUGAGCUUAAUUGGUUUGCCUUGCCAACGAGCUGGGCGGCAUUUUCCGAUUAAUUGAUGGGUGGGAAAAGUGUGU_____GGACGAGACGAAAGUCAAGUUGGCAUUUAAA .....((.(((.(((((..(((((.......))))).))))).))).(((((((((.(((........))).)).))))))).......))....(((....)))............... (-28.64 = -28.92 + 0.28)



| Location | 16,891,185 – 16,891,299 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.80 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -25.44 |
| Energy contribution | -26.56 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16891185 114 + 22224390 ACUUUUCCCACCCAUCAAUUAAUCGGAAAAUGCCGCCCAGCUCGUUGGCAAGGCAAACCAAUUAAGCUCAGCCGGUUAAGCCAAUGGCUUUGGUAAACAUUCUGGGAUAUGGGG------ ......((((((((....((((((((....((((..((((....))))...))))................))))))))(((((.....)))))........))))...)))).------ ( -31.45) >DroSec_CAF1 90156 116 + 1 ACUUUUCCCACCCAUCAAUUAAUCGGAAAAUGCCGCCCAGCUCGUUGGCAAGGCAAACCAAUUAAGCUCAGCCGGUUAAGCCAAUAGCUUUGGUAAACAUACCGGGAUAUGGGAUA---- .....(((((...(((..(((((.((....((((..((((....))))...))))..)).)))))......(((((...(((((.....)))))......)))))))).)))))..---- ( -33.10) >DroSim_CAF1 70603 120 + 1 ACUUUUCCCACCCAUCAAUUAAUCGGAAAAUACCGCCCAGCUCGUUGGCAAGGCAAACCAAUUAAGCUCAGCCGGUUAAGCCAAUAGCUUUGGUAAACAUACCGGGAUAUGGGAUAUGCU .....(((((.............(((......)))(((((((.((((((..((....))..((((.(......).))))))))))))))..((((....)))))))...)))))...... ( -32.00) >DroEre_CAF1 74648 113 + 1 ACUUUUCCCACCCAUCAAUUAAUCGGAAAAUGCCGCCCAGCUCGUCGGCAAGGCAAACCAAUUAAGCUCAGCCGGUUAAGCCAAUAGCUUUGGUAAACAUUUCGGGU-------UUUGGG ......(((((((.....((((((((....(((((.(......).))))).((....))............))))))))(((((.....))))).........))))-------...))) ( -30.60) >DroYak_CAF1 76628 120 + 1 ACUUUUCCCACCCAUCAGUUAAUCGGAAAAUGCCGCCCAGCUCGUUGGCAAGGCAAACCAAUUAAGCUCAGCCGGUUAAGCCAAUAGCUUUGGUAAACAUUCCGGGAAAUGCGAUAUGGU .........(((.(((.(((..((((((..((((..((((....))))...)))).(((((...((((.....((.....))...))))))))).....))))))..)))..)))..))) ( -30.10) >consensus ACUUUUCCCACCCAUCAAUUAAUCGGAAAAUGCCGCCCAGCUCGUUGGCAAGGCAAACCAAUUAAGCUCAGCCGGUUAAGCCAAUAGCUUUGGUAAACAUUCCGGGAUAUGGGAUAUG__ .....((((((((.....((((((((....(((((.(......).))))).((....))............))))))))(((((.....))))).........)))...)))))...... (-25.44 = -26.56 + 1.12)

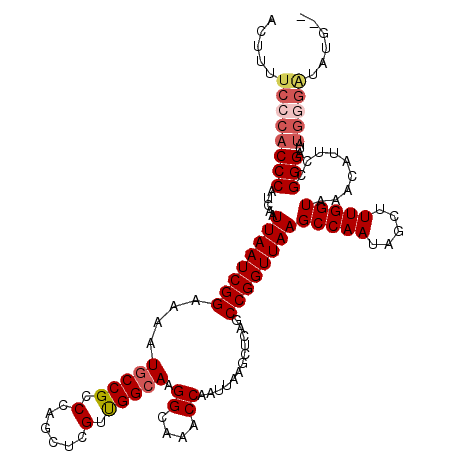

| Location | 16,891,185 – 16,891,299 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.80 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -30.32 |
| Energy contribution | -30.28 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16891185 114 - 22224390 ------CCCCAUAUCCCAGAAUGUUUACCAAAGCCAUUGGCUUAACCGGCUGAGCUUAAUUGGUUUGCCUUGCCAACGAGCUGGGCGGCAUUUUCCGAUUAAUUGAUGGGUGGGAAAAGU ------.((((...((((....(((((((.(((((...)))))....)).)))))((((((((..((((..(((.........)))))))....))))))))....))))))))...... ( -35.20) >DroSec_CAF1 90156 116 - 1 ----UAUCCCAUAUCCCGGUAUGUUUACCAAAGCUAUUGGCUUAACCGGCUGAGCUUAAUUGGUUUGCCUUGCCAACGAGCUGGGCGGCAUUUUCCGAUUAAUUGAUGGGUGGGAAAAGU ----..(((((...(((((((....)))).(((((...((((.....)))).)))))((((((..((((..(((.........)))))))....)))))).......))))))))..... ( -37.50) >DroSim_CAF1 70603 120 - 1 AGCAUAUCCCAUAUCCCGGUAUGUUUACCAAAGCUAUUGGCUUAACCGGCUGAGCUUAAUUGGUUUGCCUUGCCAACGAGCUGGGCGGUAUUUUCCGAUUAAUUGAUGGGUGGGAAAAGU ......(((((...(((((((....))))......(((((....((((.((.(((((..(((((.......))))).))))).)))))).....)))))........))))))))..... ( -38.10) >DroEre_CAF1 74648 113 - 1 CCCAAA-------ACCCGAAAUGUUUACCAAAGCUAUUGGCUUAACCGGCUGAGCUUAAUUGGUUUGCCUUGCCGACGAGCUGGGCGGCAUUUUCCGAUUAAUUGAUGGGUGGGAAAAGU (((...-------(((((....(((((((.((((.....))))....)).)))))((((((((..((((..(((.(.....).)))))))....))))))))....))))))))...... ( -33.50) >DroYak_CAF1 76628 120 - 1 ACCAUAUCGCAUUUCCCGGAAUGUUUACCAAAGCUAUUGGCUUAACCGGCUGAGCUUAAUUGGUUUGCCUUGCCAACGAGCUGGGCGGCAUUUUCCGAUUAACUGAUGGGUGGGAAAAGU ........((.((((((((((((((..(((..(((.(((((......(((.(((((.....))))))))..)))))..))))))..))))))))((.(((....))).)).)))))).)) ( -35.70) >consensus __CAUAUCCCAUAUCCCGGAAUGUUUACCAAAGCUAUUGGCUUAACCGGCUGAGCUUAAUUGGUUUGCCUUGCCAACGAGCUGGGCGGCAUUUUCCGAUUAAUUGAUGGGUGGGAAAAGU .............((((((((((((.....((((.....)))).....(((.(((((..(((((.......))))).))))).)))))))))))((.(((....))).)).))))..... (-30.32 = -30.28 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:57 2006