
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,871,455 – 16,871,581 |
| Length | 126 |
| Max. P | 0.703611 |

| Location | 16,871,455 – 16,871,551 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 85.48 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.72 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
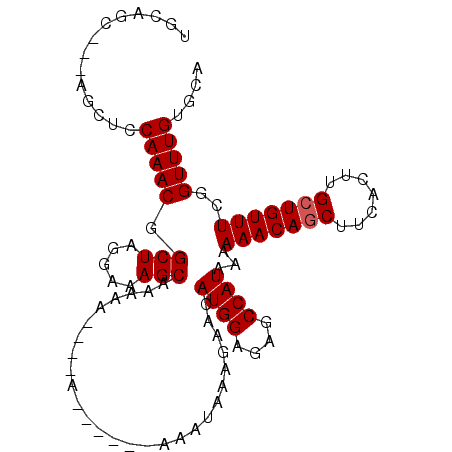
>X_DroMel_CAF1 16871455 96 + 22224390 UGCAGC---AGCUCCAAACGGCUAGGAAAGCAAAAAAAAAA------AAAUAAAGAACAUGGAGAGCCAUAAAAAACAGCUUCACUUGGUGUUUCGGUUUGUGCA .(((.(---((..(((((((.(.((..((((..........------...(....)..((((....))))........))))..)).).))))).)).)))))). ( -17.50) >DroSec_CAF1 69931 91 + 1 UGCAGC---AGCUCCAAACGGCUAGGAAAGCAAAA-----A------AAAUAAAGAACAUGGAGAGCCAUAAAAAACAGCUUCACUUGCUGUUUCGGUUUGUGCA .(((.(---((..(((((((((.((..((((....-----.------...(....)..((((....))))........))))..)).))))))).)).)))))). ( -22.00) >DroSim_CAF1 50432 92 + 1 NNNNNN---NNNUCCAAACGGCUAGGAAAGCAAAAA----A------AAAUAAAGAACAUGGAGAGCCAUAAAAAACAGCUUCACUUGCUGUUUCGGUUUGUGCA ......---....(((((((((.((..((((.....----.------...(....)..((((....))))........))))..)).))))))).))........ ( -16.50) >DroYak_CAF1 56540 105 + 1 UGCUGCAGCAGCUCCAAACGGCUAGAAAAGCAAAAAAAAAACAAAAAGAGUAAAGAACAUGGAGAGCCAUAAAAAACAGCUUCACUUGCUGUUUCGGUUUGUGCA ....(((.(((..(((((((((.((..((((..................((.....))((((....))))........))))..)).))))))).)).)))))). ( -23.40) >consensus UGCAGC___AGCUCCAAACGGCUAGGAAAGCAAAAA____A______AAAUAAAGAACAUGGAGAGCCAUAAAAAACAGCUUCACUUGCUGUUUCGGUUUGUGCA ..............(((((.(((.....)))...........................((((....))))...(((((((.......)))))))..))))).... (-14.48 = -14.72 + 0.25)


| Location | 16,871,484 – 16,871,581 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 89.60 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -17.83 |
| Energy contribution | -18.57 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
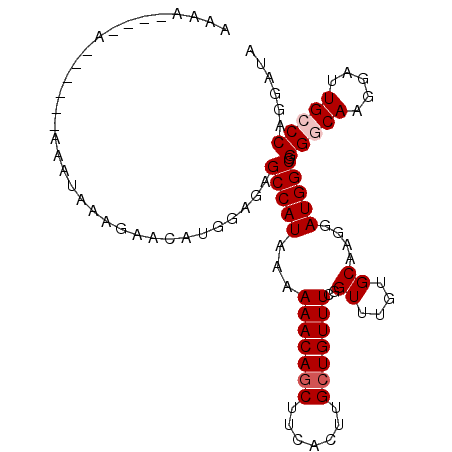
>X_DroMel_CAF1 16871484 97 + 22224390 AAAAAAAAA------AAAUAAAGAACAUGGAGAGCCAUAAAAAACAGCUUCACUUGGUGUUUCGGUUUGUGCAAGGAUGGCGGGACAAGGAUUGCCCAGGCUU .........------................(((((..........(((...(((.(..(........)..))))...)))(((.((.....))))).))))) ( -18.70) >DroSec_CAF1 69960 92 + 1 AAA-----A------AAAUAAAGAACAUGGAGAGCCAUAAAAAACAGCUUCACUUGCUGUUUCGGUUUGUGCAAGGAUGGCGGGGCAAGAAUUGCCCAGGAUA ...-----.------..................(((((...(((((((.......)))))))..((....))....))))).(((((.....)))))...... ( -23.30) >DroSim_CAF1 50461 93 + 1 AAAA----A------AAAUAAAGAACAUGGAGAGCCAUAAAAAACAGCUUCACUUGCUGUUUCGGUUUGUGCAAGGAUGGCGGGGCAAGGAUUGCCCAGGAUA ....----.------..................(((((...(((((((.......)))))))..((....))....))))).(((((.....)))))...... ( -23.30) >DroYak_CAF1 56572 103 + 1 AAAAAAAAACAAAAAGAGUAAAGAACAUGGAGAGCCAUAAAAAACAGCUUCACUUGCUGUUUCGGUUUGUGCAAGGAUGGCGGGGCAAGGAUUGGCCAGGAUU ........(((((..((.........((((....))))....((((((.......))))))))..))))).......((((.((.......)).))))..... ( -20.40) >consensus AAAA____A______AAAUAAAGAACAUGGAGAGCCAUAAAAAACAGCUUCACUUGCUGUUUCGGUUUGUGCAAGGAUGGCGGGGCAAGGAUUGCCCAGGAUA .................................(((((...(((((((.......)))))))..((....))....))))).(((((.....)))))...... (-17.83 = -18.57 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:51 2006