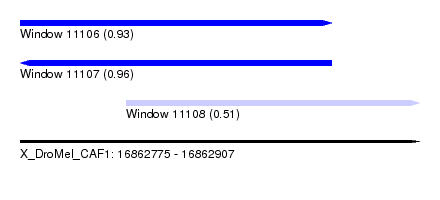
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,862,775 – 16,862,907 |
| Length | 132 |
| Max. P | 0.958093 |

| Location | 16,862,775 – 16,862,878 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.87 |
| Mean single sequence MFE | -32.07 |
| Consensus MFE | -28.54 |
| Energy contribution | -29.43 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
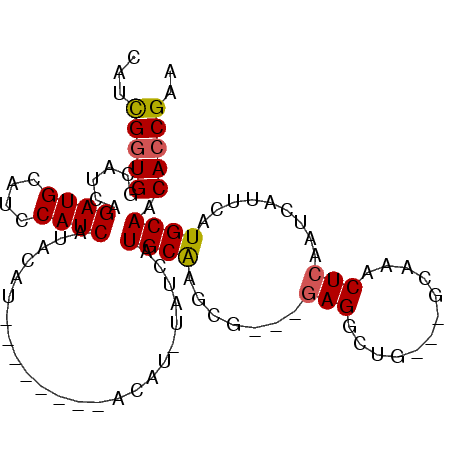
>X_DroMel_CAF1 16862775 103 + 22224390 UGAAGA-AGGCCCAGCAAAUGGGGCCGAGCCACCGAUAUCGUUGGCAUCAGAUGCAUCCAUCAUACAUACAUACAUACAUGUAUCAUGCAAGCAAGCAAGGCUG- ......-.(((((........)))))((((((.((....)).)))).))...(((...(((.((((((..........)))))).)))...)))(((...))).- ( -27.80) >DroSec_CAF1 61404 94 + 1 UGGAGAUGGGCCCAGCAAAUGGGGCCUAGCCACCGACAUUGGUGGCAUCAGAUGCAUCCAUCAUACAU--------ACAUAUAUCAUGCAAGCG---GAGGCUGG ((((..(((((((........)))))))(((((((....)))))))..........))))........--------..............(((.---...))).. ( -31.00) >DroSim_CAF1 41438 94 + 1 UGGAGAUGGGCCCAGCAAAUGGGGCCUAGCCACCGACAUCGGUGGCAUCAGAUGCAUACAUCAUACAU--------ACAUGUAUCAUGCAAGCG---GAGGCUGG ......(((((((........)))))))(((((((....)))))))..(((.(((...(((.(((((.--------...))))).)))...)))---....))). ( -37.40) >consensus UGGAGAUGGGCCCAGCAAAUGGGGCCUAGCCACCGACAUCGGUGGCAUCAGAUGCAUCCAUCAUACAU________ACAUGUAUCAUGCAAGCG___GAGGCUGG (((...(((((((........)))))))(((((((....))))))).)))..(((((.....((((((..........)))))).)))))(((.......))).. (-28.54 = -29.43 + 0.89)


| Location | 16,862,775 – 16,862,878 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.87 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -29.12 |
| Energy contribution | -29.57 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16862775 103 - 22224390 -CAGCCUUGCUUGCUUGCAUGAUACAUGUAUGUAUGUAUGUAUGAUGGAUGCAUCUGAUGCCAACGAUAUCGGUGGCUCGGCCCCAUUUGCUGGGCCU-UCUUCA -..((((.((.(((.(.(((.(((((((........))))))).))).).))).((((.((((.((....)).))))))))........)).))))..-...... ( -32.90) >DroSec_CAF1 61404 94 - 1 CCAGCCUC---CGCUUGCAUGAUAUAUGU--------AUGUAUGAUGGAUGCAUCUGAUGCCACCAAUGUCGGUGGCUAGGCCCCAUUUGCUGGGCCCAUCUCCA ...((.((---(((.((((((.......)--------))))).).)))).))....(((((((((......))))))..(((((........))))).))).... ( -32.70) >DroSim_CAF1 41438 94 - 1 CCAGCCUC---CGCUUGCAUGAUACAUGU--------AUGUAUGAUGUAUGCAUCUGAUGCCACCGAUGUCGGUGGCUAGGCCCCAUUUGCUGGGCCCAUCUCCA ........---.((.(((((.(((((...--------.))))).))))).))....((((((((((....)))))))..(((((........))))).))).... ( -37.30) >consensus CCAGCCUC___CGCUUGCAUGAUACAUGU________AUGUAUGAUGGAUGCAUCUGAUGCCACCGAUGUCGGUGGCUAGGCCCCAUUUGCUGGGCCCAUCUCCA ............((.(.(((.((((((..........)))))).))).).)).......(((((((....)))))))..(((((........)))))........ (-29.12 = -29.57 + 0.45)



| Location | 16,862,810 – 16,862,907 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 75.90 |
| Mean single sequence MFE | -20.91 |
| Consensus MFE | -12.51 |
| Energy contribution | -12.59 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16862810 97 + 22224390 UAUCGUUGGCAUCAGAUGCAUCCAUCAUACAUACAUACAUACAUGUAUCAUGCAAGCAAGCAAGGCUG---GCAAGCUCAAUCAUUCAUGCAACACCGAA ....((((((((..((((....))))....((((((......)))))).))))..((.(((...))).---)).................))))...... ( -16.90) >DroSec_CAF1 61440 89 + 1 CAUUGGUGGCAUCAGAUGCAUCCAUCAUACAU--------ACAUAUAUCAUGCAAGCG---GAGGCUGGAGGCAAACUCAAUCAUUCAUGCAACACCGAA ..((((((......((((....))))......--------..........(((((((.---...))).(((.....))).........)))).)))))). ( -19.90) >DroSim_CAF1 41474 89 + 1 CAUCGGUGGCAUCAGAUGCAUACAUCAUACAU--------ACAUGUAUCAUGCAAGCG---GAGGCUGGAGGCAAACUCAAUCAUUCAUGCAACACCGAA ..((((((((((..(((((((...........--------..)))))))))))..(((---(((....(((.....))).....))).)))..)))))). ( -26.82) >DroEre_CAF1 46518 74 + 1 AAACGGUGCCACCAGAUGCAUCCAUCGAGC--------------------UGCGGGCG---GAGGAAG---GCAAACUCAAUCAUUCAUGCAACACCGAA ...(((((((.((.((((....))))..((--------------------.....)))---).))...---(((..............)))..))))).. ( -17.54) >DroYak_CAF1 46210 74 + 1 CAACGGUGGCAUCGGAUGCAUCCAUCAUCC--------------------UGCAGGCG---GAGGAUG---GCAAACUCAAUCAUUCAUGCAACACCGAA ...(((((((((.(((((......((((((--------------------(.(....)---.))))))---)..........)))))))))..))))).. ( -23.39) >consensus CAUCGGUGGCAUCAGAUGCAUCCAUCAUACAU________ACAU_UAUCAUGCAAGCG___GAGGCUG___GCAAACUCAAUCAUUCAUGCAACACCGAA ...(((((......((((....))))........................((((.......(((............))).........)))).))))).. (-12.51 = -12.59 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:49 2006