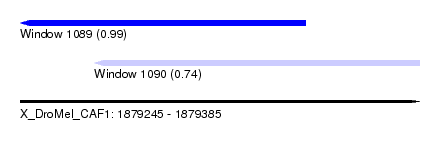
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,879,245 – 1,879,385 |
| Length | 140 |
| Max. P | 0.992719 |

| Location | 1,879,245 – 1,879,345 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 90.19 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.56 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1879245 100 - 22224390 CUGCGGGAUCCUGUCUUCCCUUUUCAGUUCAGCAGCAGCCUAAGGUUGGAUUAUAAUCUUGGCCUCAAUU-UU---UUUGUGCUGCAUUCUCUCUUAAAUUACU ....((((........))))...........(((((((((.(((((((.....))))))))))..(((..-..---.))))))))).................. ( -25.20) >DroSec_CAF1 4015 100 - 1 CUGCGGGAUCCUGUCUUCCCU-UUCAGUUCAGCAGCAGCCUAAGGUUGGAUUAUAAUCUUGGCCAAAAUUAUU---UUUGUGCUGCAUUCUUUCUUAAAUUACU (((.((((........)))).-..)))....(((((((((.(((((((.....))))))))))(((((....)---)))))))))).................. ( -27.80) >DroSim_CAF1 5418 100 - 1 CUGCGGGAUCCUGUCUUCCCU-UUCAGUUCAGCAGCAGCCUAAGGUUGGAUUAUAAUCUUGGCCAAAAUUCUU---UUUGUGCUGCAUUCUCUCUUAAAUUACU (((.((((........)))).-..)))....(((((((((.(((((((.....))))))))))(((((....)---)))))))))).................. ( -27.80) >DroEre_CAF1 4070 100 - 1 CUCCGGGAUCCUGGCUUCCCU-UUCAGUUCAGCAGCAGCAUAAGGUUGGAUUAUGAUCUUGGCCACAAUUUCUUGUUUUGUGCUGCAUU---UCUUAAAGUAGU ....((((........)))).-............((((((((((((..(.........)..)))((((....)))).)))))))))...---............ ( -24.70) >DroYak_CAF1 4653 101 - 1 CUCCGGGAUCCUGGCUUCCCUUUUCAGUUCAGCAGCAGCCUAAGGUUCGUUUAUGAUCUUGGCCACAAUUUCU---UUUGUGCUGCAUUCUCUCUUAAAGUAGU ....((((........))))...........(((((.(((.((((((.......))))))))).((((.....---.))))))))).................. ( -24.70) >consensus CUGCGGGAUCCUGUCUUCCCU_UUCAGUUCAGCAGCAGCCUAAGGUUGGAUUAUAAUCUUGGCCACAAUUUUU___UUUGUGCUGCAUUCUCUCUUAAAUUACU ....((((........))))...........(((((((((.(((((((.....)))))))))).................)))))).................. (-21.40 = -21.56 + 0.16)

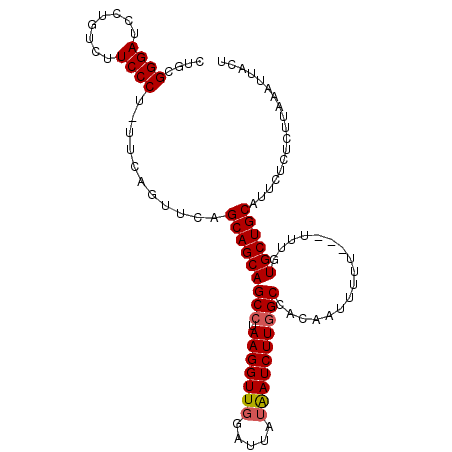
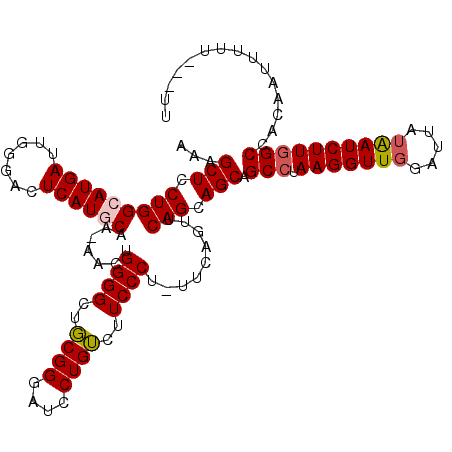
| Location | 1,879,271 – 1,879,385 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 91.44 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -30.72 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1879271 114 - 22224390 AAAGCUCCUGGCAUGAUUGGAGCUCAUGCAAAAACUGGGGCUGCGGGAUCCUGUCUUCCCUUUUCAGUUCAGCAGCAGCCUAAGGUUGGAUUAUAAUCUUGGCCUCAAUU-UU---UU ...(((.(((((((((.......))))))...(((((..(....((((........)))).)..)))))))).))).(((.(((((((.....)))))))))).......-..---.. ( -34.80) >DroSec_CAF1 4041 113 - 1 AAAGCUCCUGGCAUGAUUAGGACUCAUGCAA-AACUGGGGCUGCGGGAUCCUGUCUUCCCU-UUCAGUUCAGCAGCAGCCUAAGGUUGGAUUAUAAUCUUGGCCAAAAUUAUU---UU ...(((.(((((((((.......))))))..-(((((..(....((((........)))))-..)))))))).))).(((.(((((((.....))))))))))..........---.. ( -34.70) >DroSim_CAF1 5444 113 - 1 AAAGCUCCUGGCAUGAUUAGGACUCAUGCAA-AACUGGGGCUGCGGGAUCCUGUCUUCCCU-UUCAGUUCAGCAGCAGCCUAAGGUUGGAUUAUAAUCUUGGCCAAAAUUCUU---UU ...(((.(((((((((.......))))))..-(((((..(....((((........)))))-..)))))))).))).(((.(((((((.....))))))))))..........---.. ( -34.70) >DroEre_CAF1 4093 116 - 1 AAAGCUCCUGGGAUGAUUGGGACUCAUGCAA-AACUGGGGCUCCGGGAUCCUGGCUUCCCU-UUCAGUUCAGCAGCAGCAUAAGGUUGGAUUAUGAUCUUGGCCACAAUUUCUUGUUU ...(((.(((((.(((..((((((((.....-...))))...((((....))))..)))).-.))).))))).))).((.((((((((.....))))))))))............... ( -32.50) >DroYak_CAF1 4679 115 - 1 AAAGCUCCUGGGAUGAUUGGGACUCAUGCAAAAACUGGGGCUCCGGGAUCCUGGCUUCCCUUUUCAGUUCAGCAGCAGCCUAAGGUUCGUUUAUGAUCUUGGCCACAAUUUCU---UU ...(((.(((((.(((..((((((((.(......)))))...((((....))))..))))...))).))))).))).(((.((((((.......)))))))))..........---.. ( -31.70) >consensus AAAGCUCCUGGCAUGAUUGGGACUCAUGCAA_AACUGGGGCUGCGGGAUCCUGUCUUCCCU_UUCAGUUCAGCAGCAGCCUAAGGUUGGAUUAUAAUCUUGGCCACAAUUUUU___UU ...(((.(((((((((.......)))))).......((((..((((....))))..)))).........))).))).(((.(((((((.....))))))))))............... (-30.72 = -30.80 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:56 2006