
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,853,895 – 16,854,078 |
| Length | 183 |
| Max. P | 0.969060 |

| Location | 16,853,895 – 16,853,999 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.32 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -25.78 |
| Energy contribution | -24.86 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.904107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16853895 104 - 22224390 CCGCAUAAAUCAUUUCUCCUCGAAGGGAGAAGCGUUGUGCAGCGUUGCACAAUGUUGUGUGUGGUUUUUCCAC--UGCAUUCGGAAAAUGGCAUUGAAAAGCUGUU (((((((.......((((((....))))))((((((((((......)))))))))).))))))).((((((..--.......)))))).(((........)))... ( -34.70) >DroSec_CAF1 52505 103 - 1 CCGCAUAAAUCAUUUCUCCUCGGAGGGUUAAGCAC-ACGCAGCGUUGCACAAUGUUGUGUGUGGUUUUUCCAC--UGCAUUCGGAAAAUGGCAUUGAAAAGCUGUU ..(((.....(((((.(((.(((.(((..((.(((-((((((((((....))))))))))))).))..))).)--)).....))))))))((........))))). ( -31.80) >DroSim_CAF1 36943 103 - 1 CCGCAUAAAUCAUUUCUCCUCGGAGGGUUAAGCAU-ACGCAGCGUUGCACAAUGUUGUGUGUGGUUUUUCCAC--UGCAUUCGGAAAAUGGCAUUGAAAAGCUGUU ..(((.....(((((.(((.(((.(((..((.(((-((((((((((....))))))))))))).))..))).)--)).....))))))))((........))))). ( -29.90) >DroEre_CAF1 37867 103 - 1 CCGCAUAAAUCAUUUCUCCUUGGAGGGUUAAGCAU-ACGCAGCGUUGCACAGUGCUGUGUGUGGUUUUUCCAC--UGCAUUUGGAAAAUGGCAUUGAAAAGCUGUU ..(((.....(((((.(((.((.((((..((.(((-((((((((((....))))))))))))).))...)).)--).))...))))))))((........))))). ( -30.60) >DroYak_CAF1 37281 105 - 1 CCGCAUAAAUCAUUUCUCCUUGGAGAGUUAAGCAU-ACGCAGCGUUGCACCGCGUUGUGUGCGGUUUUUCCACAUUGCAUUCGGAAAACGGCAUUGAAAAGCUGUU ((((.........(((((....)))))........-(((((((((......)))))))))))))..(((((...........)))))(((((........))))). ( -31.00) >consensus CCGCAUAAAUCAUUUCUCCUCGGAGGGUUAAGCAU_ACGCAGCGUUGCACAAUGUUGUGUGUGGUUUUUCCAC__UGCAUUCGGAAAAUGGCAUUGAAAAGCUGUU (((..........(((((....)))))....(((..((((((((((....))))))))))((((.....))))..)))...)))..((((((........)))))) (-25.78 = -24.86 + -0.92)


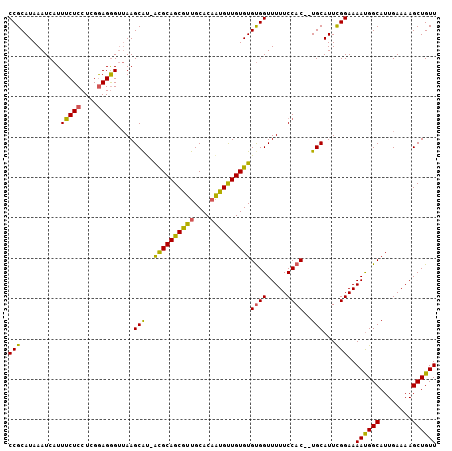
| Location | 16,853,929 – 16,854,039 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 90.04 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -25.37 |
| Energy contribution | -25.45 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16853929 110 - 22224390 UUCUACCGUUUCCCAUUUUUUCCGCGCUUUUCCUCGUGCACCGCAUAAAUCAUUUCUCCUCGAAGGGAGAAGCGUUGUGCAGCGUUGCACAAUGUUGUGUGUGGUUUUUC .......................((((........))))((((((((.......((((((....))))))((((((((((......)))))))))).))))))))..... ( -35.70) >DroSec_CAF1 52539 109 - 1 UUCCACCGUUUCCAAUUUUUUCCGCGCUUUUCUUCGUGCACCGCAUAAAUCAUUUCUCCUCGGAGGGUUAAGCAC-ACGCAGCGUUGCACAAUGUUGUGUGUGGUUUUUC ....(((................((((........))))................(((....)))))).((.(((-((((((((((....))))))))))))).)).... ( -29.50) >DroSim_CAF1 36977 109 - 1 UUCCACCGUUUCCCAUUUUUUCCGCGCUUUUCUUCGUGCACCGCAUAAAUCAUUUCUCCUCGGAGGGUUAAGCAU-ACGCAGCGUUGCACAAUGUUGUGUGUGGUUUUUC ....((((((((((.........((((........)))).(((.................))).)))..))))((-((((((((((....)))))))))))))))..... ( -28.43) >DroEre_CAF1 37901 107 - 1 UUCUACCGUUU-CCCGUUUUUCCGCGCUU-UCUUCUUGCACCGCAUAAAUCAUUUCUCCUUGGAGGGUUAAGCAU-ACGCAGCGUUGCACAGUGCUGUGUGUGGUUUUUC ....(((....-...((((..(((((...-...........)))...........(((....)))))..))))((-((((((((((....)))))))))))))))..... ( -26.24) >DroYak_CAF1 37317 109 - 1 UUCCACCGUUGGCCAGUUUUUCCGCGCUUUUCCUCGUGCACCGCAUAAAUCAUUUCUCCUUGGAGAGUUAAGCAU-ACGCAGCGUUGCACCGCGUUGUGUGCGGUUUUUC ..(((....)))...........((((........))))(((((.........(((((....)))))........-(((((((((......))))))))))))))..... ( -30.30) >consensus UUCCACCGUUUCCCAUUUUUUCCGCGCUUUUCUUCGUGCACCGCAUAAAUCAUUUCUCCUCGGAGGGUUAAGCAU_ACGCAGCGUUGCACAAUGUUGUGUGUGGUUUUUC .....................((((((........(((((.(((.........(((((....)))))....((.....)).))).)))))........))))))...... (-25.37 = -25.45 + 0.08)

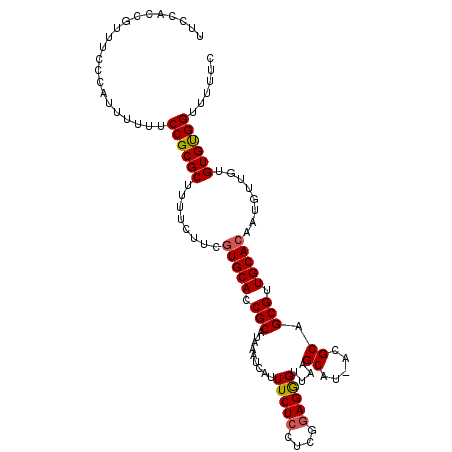
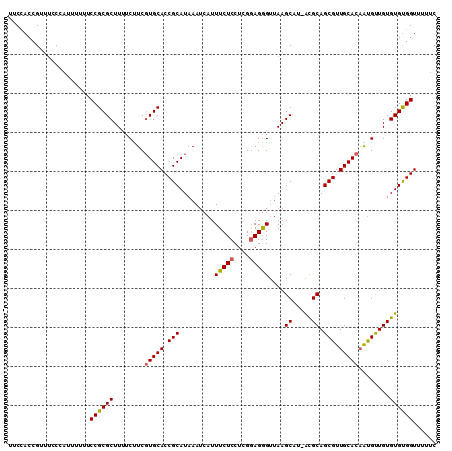
| Location | 16,853,963 – 16,854,078 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.10 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16853963 115 + 22224390 AACGCUUCUCCCUUCGAGGAGAAAUGAUUUAUGCGGUGCACGAGGAAAAGCGCGGAAAAAAUGGGAAACGGUAGAAAACGAUG-AAAGCAUAAAAGAGAAAGUGGAAAGUGAUGAA ..(((((((((......))))))....(((((((.((((..........)))).......((.(....).))...........-...))))))).......)))............ ( -23.90) >DroSec_CAF1 52573 114 + 1 -GUGCUUAACCCUCCGAGGAGAAAUGAUUUAUGCGGUGCACGAAGAAAAGCGCGGAAAAAAUUGGAAACGGUGGAAAACGAUG-CAAGCAUAAAAGAGAAAGUGGAAAGUGAUGAA -..((((..(((((....)))......(((((((..((((((........))((......((((....))))......)).))-)).))))))).........)).))))...... ( -22.40) >DroSim_CAF1 37011 114 + 1 -AUGCUUAACCCUCCGAGGAGAAAUGAUUUAUGCGGUGCACGAAGAAAAGCGCGGAAAAAAUGGGAAACGGUGGAAAGCGAUG-CAAGCAUAAAAGAGAAAGUGGAAAGUGAUGAA -..((((..(((((....)))......(((((((..((((..........(((.......((.(....).)).....))).))-)).))))))).........)).))))...... ( -22.70) >DroYak_CAF1 37351 113 + 1 -AUGCUUAACUCUCCAAGGAGAAAUGAUUUAUGCGGUGCACGAGGAAAAGCGCGGAAAAACUGGCCAACGGUGGAAAAUGAUAGAAAGCAUAUGAGAGAAAGUGGAAAGUGAA--G -...(((.(((.((((.............(((((.((((..........))))......((((.....))))...............)))))..........)))).))).))--) ( -19.60) >consensus _AUGCUUAACCCUCCGAGGAGAAAUGAUUUAUGCGGUGCACGAAGAAAAGCGCGGAAAAAAUGGGAAACGGUGGAAAACGAUG_AAAGCAUAAAAGAGAAAGUGGAAAGUGAUGAA ..(((((..(.(((....)))......(((((((.((((..........))))..........(....)..................))))))).........)..)))))..... (-17.41 = -17.10 + -0.31)

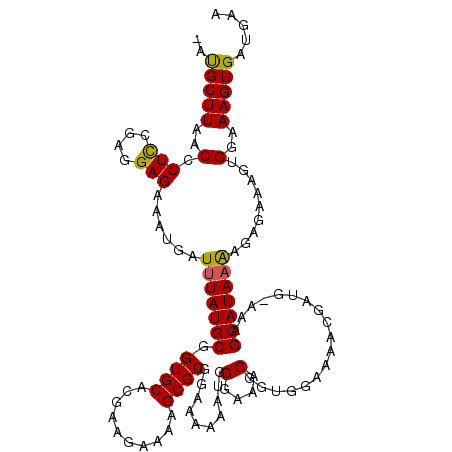
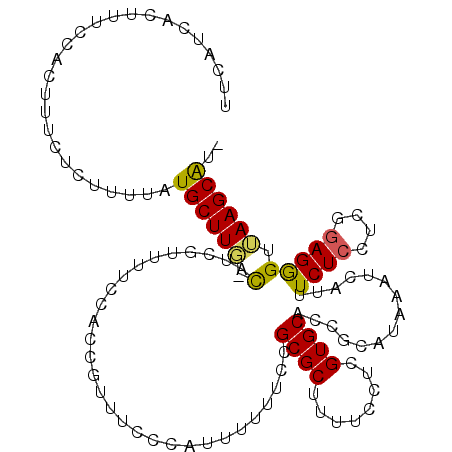
| Location | 16,853,963 – 16,854,078 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -18.28 |
| Consensus MFE | -13.19 |
| Energy contribution | -12.88 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16853963 115 - 22224390 UUCAUCACUUUCCACUUUCUCUUUUAUGCUUU-CAUCGUUUUCUACCGUUUCCCAUUUUUUCCGCGCUUUUCCUCGUGCACCGCAUAAAUCAUUUCUCCUCGAAGGGAGAAGCGUU ......................(((((((...-....((.....)).................((((........))))...)))))))...((((((((....)))))))).... ( -19.20) >DroSec_CAF1 52573 114 - 1 UUCAUCACUUUCCACUUUCUCUUUUAUGCUUG-CAUCGUUUUCCACCGUUUCCAAUUUUUUCCGCGCUUUUCUUCGUGCACCGCAUAAAUCAUUUCUCCUCGGAGGGUUAAGCAC- ......(((((((.........(((((((.((-((.((........(((..............)))........))))))..)))))))............))))))).......- ( -17.13) >DroSim_CAF1 37011 114 - 1 UUCAUCACUUUCCACUUUCUCUUUUAUGCUUG-CAUCGCUUUCCACCGUUUCCCAUUUUUUCCGCGCUUUUCUUCGUGCACCGCAUAAAUCAUUUCUCCUCGGAGGGUUAAGCAU- .........................(((((((-....(((((((...(.....).........((((........))))......................))))))))))))))- ( -17.00) >DroYak_CAF1 37351 113 - 1 C--UUCACUUUCCACUUUCUCUCAUAUGCUUUCUAUCAUUUUCCACCGUUGGCCAGUUUUUCCGCGCUUUUCCUCGUGCACCGCAUAAAUCAUUUCUCCUUGGAGAGUUAAGCAU- (--((.((((((((..........(((((.............(((....)))...........((((........))))...))))).............)))))))).)))...- ( -19.80) >consensus UUCAUCACUUUCCACUUUCUCUUUUAUGCUUG_CAUCGUUUUCCACCGUUUCCCAUUUUUUCCGCGCUUUUCCUCGUGCACCGCAUAAAUCAUUUCUCCUCGGAGGGUUAAGCAU_ ..........................((((((.(.............................((((........))))...............((((....))))).)))))).. (-13.19 = -12.88 + -0.31)


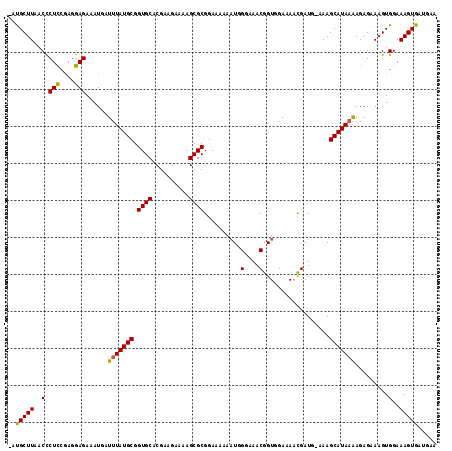
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:42 2006