
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,822,227 – 16,822,322 |
| Length | 95 |
| Max. P | 0.906041 |

| Location | 16,822,227 – 16,822,322 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.52 |
| Mean single sequence MFE | -18.65 |
| Consensus MFE | -10.02 |
| Energy contribution | -11.38 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
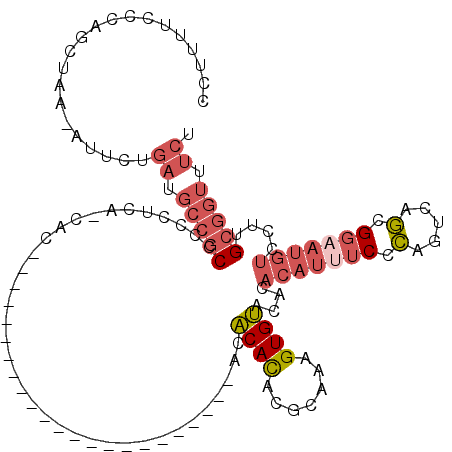
>X_DroMel_CAF1 16822227 95 - 22224390 CCUUUUCCCAGCUAA-AUUCUGAUGCCGCCCCCUUA-CAC-----------------------ACACAUACGCAAAGUGUACACACAUUUCCCAGUCAGCGGAAUGUCCUUGCGGUUUCU ...............-.....((.(((((.....((-(((-----------------------.............)))))...(((((((.(.....).)))))))....))))).)). ( -16.82) >DroSec_CAF1 19940 96 - 1 CCUUUUCCCAGCUAA-AUUCUGAUGCCGCCCCCUCAUCAC-----------------------ACACACACGCCAAGUGUACACACAUUUCCCAGUCAGCGGAAUGUCCUUGCGGUUUCU ...............-.....((.(((((...........-----------------------....((((.....))))....(((((((.(.....).)))))))....))))).)). ( -17.70) >DroSim_CAF1 4688 96 - 1 CCUUUUCCCAGCUAA-AUUCUGAUGCCGCCCCCUCAUCAC-----------------------ACACACACGCAAAGUGUACACACAUUUCCCAGUCAGCGGAAUGUCCUUGCGGUUUCU ...............-.....((.(((((...........-----------------------....((((.....))))....(((((((.(.....).)))))))....))))).)). ( -17.70) >DroEre_CAF1 4650 105 - 1 CCAUUUCCCAGCUAA-AUUCUGAAGCCGCCCC-UCGCCACA-------------CACACAAAUACACAUACGUAAAGUGUACACACAUCUCACAGUCAGCGGAAUGUCCUUGCGGUUUCU ...............-.....((((((((...-........-------------.((((...(((......)))..))))....((((..(.(.....).)..))))....)))))))). ( -19.20) >DroWil_CAF1 5524 74 - 1 -------CCCACUAAC---CAUUUGGCGCUUCUUCA-CUC-----------------------ACUCACACACUGAGUGACUA------ACUGAGUGACCGAGCUCUCUU-G-A----CA -------.........---..((((((((((.....-.((-----------------------(((((.....)))))))...------...))))).))))).......-.-.----.. ( -17.60) >DroYak_CAF1 4730 118 - 1 CCUUUUUCCAGCUAA-AUUCUGAAGCCGCCCC-UCAACACACUAAACACACUCGCACACACUCACACAUACGUAAGGUGUACACACAUUUCCCAGUCAGCGGAAUGUCCUUGCGGUUUCU ...............-.....((((((((...-....................(.((((.((.((......)).)))))).)..(((((((.(.....).)))))))....)))))))). ( -22.90) >consensus CCUUUUCCCAGCUAA_AUUCUGAUGCCGCCCCCUCA_CAC_______________________ACACACACGCAAAGUGUACACACAUUUCCCAGUCAGCGGAAUGUCCUUGCGGUUUCU .....................((.(((((....................................((((.......))))....(((((((.(.....).)))))))....))))).)). (-10.02 = -11.38 + 1.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:27 2006