
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,804,439 – 16,804,574 |
| Length | 135 |
| Max. P | 0.942673 |

| Location | 16,804,439 – 16,804,536 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 84.98 |
| Mean single sequence MFE | -18.50 |
| Consensus MFE | -15.05 |
| Energy contribution | -16.17 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16804439 97 + 22224390 CUUGGUUUAGUUCUCGCUAAAGCACACCAAUUCCCCCCACAUAUGUACAUAUAUACAUAUGUAUAAAACAUAUGUGU-CCUGAUCCAAUGCGAUCGAU ..((.((((((....)))))).)).............(((((((((((((((....)))))).....))))))))).-..(((((......))))).. ( -21.30) >DroSim_CAF1 14000 97 + 1 CUUGGUUUAGUUCUCGCUAAAGCACACCAAUUCCCCCCACAUAUGUACAUAUAUACAUAUGUAUAAAACAUAUGUGU-CCUGAUCCAAUGCGAUCGAU ..((.((((((....)))))).)).............(((((((((((((((....)))))).....))))))))).-..(((((......))))).. ( -21.30) >DroEre_CAF1 13076 81 + 1 ----GUUUAGUUCUCGCUAAAGCACACCAAAUCCC-CCGCAUAUGUAAAUA------------UAAAACAUAUGUAUACCUGAUCCAAUGCGAUCGAU ----.((((((....))))))..............-..((((((((.....------------....)))))))).....(((((......))))).. ( -12.90) >consensus CUUGGUUUAGUUCUCGCUAAAGCACACCAAUUCCCCCCACAUAUGUACAUAUAUACAUAUGUAUAAAACAUAUGUGU_CCUGAUCCAAUGCGAUCGAU ..((.((((((....)))))).)).............(((((((((...((((((....))))))..)))))))))....(((((......))))).. (-15.05 = -16.17 + 1.11)

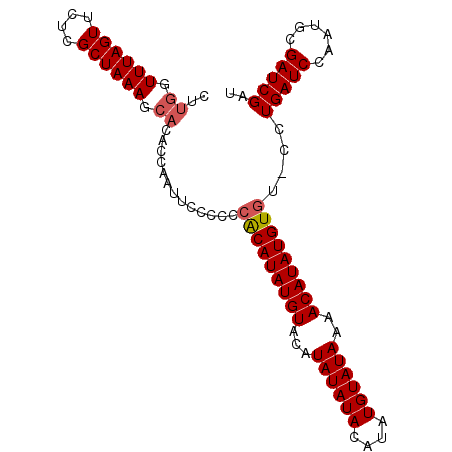
| Location | 16,804,439 – 16,804,536 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 84.98 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16804439 97 - 22224390 AUCGAUCGCAUUGGAUCAGG-ACACAUAUGUUUUAUACAUAUGUAUAUAUGUACAUAUGUGGGGGGAAUUGGUGUGCUUUAGCGAGAACUAAACCAAG ...((((......)))).((-.(((((((((..(((((....))))).....))))))))).......(((((.(((....)))...))))).))... ( -26.50) >DroSim_CAF1 14000 97 - 1 AUCGAUCGCAUUGGAUCAGG-ACACAUAUGUUUUAUACAUAUGUAUAUAUGUACAUAUGUGGGGGGAAUUGGUGUGCUUUAGCGAGAACUAAACCAAG ...((((......)))).((-.(((((((((..(((((....))))).....))))))))).......(((((.(((....)))...))))).))... ( -26.50) >DroEre_CAF1 13076 81 - 1 AUCGAUCGCAUUGGAUCAGGUAUACAUAUGUUUUA------------UAUUUACAUAUGCGG-GGGAUUUGGUGUGCUUUAGCGAGAACUAAAC---- .(((..(((((..((((.......(((((((....------------.....)))))))...-..))))..)))))......))).........---- ( -19.00) >consensus AUCGAUCGCAUUGGAUCAGG_ACACAUAUGUUUUAUACAUAUGUAUAUAUGUACAUAUGUGGGGGGAAUUGGUGUGCUUUAGCGAGAACUAAACCAAG .(((..(((((..(.((.....(((((((((..((((........))))...)))))))))....)).)..)))))......)))............. (-19.82 = -20.10 + 0.28)



| Location | 16,804,479 – 16,804,574 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -20.35 |
| Consensus MFE | -12.20 |
| Energy contribution | -14.07 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16804479 95 - 22224390 UAAAGACAAGUCCCAGGCUUGCAAAGCCCCCUAUGUAUAUCGAUCGCAUUGGAUCAGG-ACACAUAUGUUUUAUACAUAUGUAUAUAUGUACAUAU ....(.(((((.....)))))).........(((((((((.((((......))))...-..((((((((.....))))))))....))))))))). ( -24.00) >DroSim_CAF1 14040 95 - 1 UAAAGACAAGUCCCAGGCUUGCAAAGCCCCCUAUGUAUAUCGAUCGCAUUGGAUCAGG-ACACAUAUGUUUUAUACAUAUGUAUAUAUGUACAUAU ....(.(((((.....)))))).........(((((((((.((((......))))...-..((((((((.....))))))))....))))))))). ( -24.00) >DroEre_CAF1 13111 80 - 1 UAAAGACAAGUCCCAGGCUUGCAAAACCCCCUA----UAUCGAUCGCAUUGGAUCAGGUAUACAUAUGUUUUA------------UAUUUACAUAU ....(.(((((.....)))))).........((----((((((((......)))).)))))).((((((....------------.....)))))) ( -14.60) >DroYak_CAF1 12740 80 - 1 UAAAGACAAGUCCCAGGCUUGCAAAGCCCCCUA----UACCGAUCGCAUUGGAUCAGGUAAACAUAUGUCUUA------------UAUAUACAUAU ..((((((.......((((.....)))).....----((((((((......)))).))))......)))))).------------........... ( -18.80) >consensus UAAAGACAAGUCCCAGGCUUGCAAAGCCCCCUA____UAUCGAUCGCAUUGGAUCAGG_ACACAUAUGUUUUA____________UAUGUACAUAU ..((((((.......((((.....))))...(((((.((((((((......)))).)))).)))))))))))........................ (-12.20 = -14.07 + 1.88)

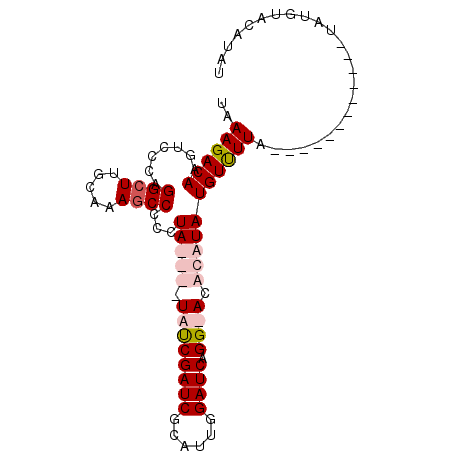

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:23 2006