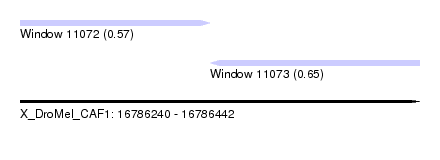
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,786,240 – 16,786,442 |
| Length | 202 |
| Max. P | 0.646109 |

| Location | 16,786,240 – 16,786,336 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.24 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -23.62 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16786240 96 + 22224390 CACCUAAGCCCCAACAGCCGCAGAUAGUGCACCAAAAUGCCCGUCUGCUGAGUGAUAGCAUGGAGAAUAUGCUGGACUCGGACCAGGAGC------------------GGGCAA .......(((((..(..(((((((..(.(((......))))..))))).((((..(((((((.....))))))).))))))....)..).------------------)))).. ( -34.30) >DroPse_CAF1 16147 105 + 1 CAAGUGGGCGCC---AGCCCCAGAUCGUGCACCAAAAUGCGCAGCUGCUGAACGACAGCCUGGAGCAUAUGCUGGACCGCGAGCAGGAGCAGUU------GGAGGCACGUGCCA ......(((((.---.(((((((...(((((......))))).(((.(((..((....((...(((....)))))....))..))).)))..))------)).)))..))))). ( -40.50) >DroSec_CAF1 4131 96 + 1 CACCUAAGCCCCAGCAGCCGCAGAUAGUGCACCAGAAUGCCCGUCUGCUGAGUGAUAGCAUGGAGAAUAUGCUGGACUCGGACCAGGAGC------------------GGGCAA .......((((..((..(((((((..(.(((......))))..)))))(((((..(((((((.....))))))).))))).....)).))------------------)))).. ( -34.90) >DroEre_CAF1 9743 114 + 1 CCCCUAAGCCCCAGCAGCCGCAGAUAGUGCACCAGAAUGCCCGUCUGCUGAGUGAUAGCAUGGAGAACAUGUUGGACUCGGAUCAGGAGCGCGCCAGAGACGAGGAGCGGGCAA .......((((..((..(((((((..(.(((......))))..)))))(((((..(((((((.....))))))).))))).....)).))((.((........)).)))))).. ( -41.00) >DroYak_CAF1 4150 114 + 1 CACCGAAGCCCCAGCUGCCGCAGAUAGUGCACCAGAAUGCCCGUCUGCUGAGUGAUAGCAUGGAGAACAUGCUGGACUCGGAUCAGGAGCGGGUCAGGGACGAGGAGAGGGCAA .......((((...((.(((((.....))).((....(((((((((.((((((..(((((((.....))))))).))))))....))).)))).)).))....)))).)))).. ( -45.10) >DroAna_CAF1 4855 114 + 1 GCCCGCAGCCGAAACAGCCGCAGAUUGUCCACCAGAAUGCCCAUCUGUUGAGCGAUAGCAUGGAGAACAUGCUGGACUCCGAUCAGGAGAGGCAACGCGACGAGGAGAGGGUCA ((((((.((.......)).))...(((((((.((((.......)))).)).(((.(((((((.....)))))))..((((.....))))......)))))))).....)))).. ( -37.20) >consensus CACCUAAGCCCCAGCAGCCGCAGAUAGUGCACCAGAAUGCCCGUCUGCUGAGUGAUAGCAUGGAGAACAUGCUGGACUCGGAUCAGGAGCGG________CGAGGAG_GGGCAA .......((((........((((((.(.(((......))).))))))).(((...(((((((.....)))))))..))).............................)))).. (-23.62 = -23.68 + 0.06)


| Location | 16,786,336 – 16,786,442 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.74 |
| Mean single sequence MFE | -41.55 |
| Consensus MFE | -18.97 |
| Energy contribution | -19.36 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16786336 106 - 22224390 GCCUUAUCGGGCACAAACCAAUUGGUGCGGUUCCUUGAGG-CACUGGCCAAGUCCACUG---GUCCACUGGC--CUUAUUUGCCGACUCCGGCAUUCGGCUGAGAUAGGGCU ((((((((((((.((......(((((.((((.((....))-.)))))))))......))---))))...(((--(.....(((((....)))))...))))..)))))))). ( -44.10) >DroSec_CAF1 4227 106 - 1 GCCUUAUCCGGCACAAACCAGUUGGUGCGGUUCCUUGAGG-CACUGGCCAAGUCCACUG---GUCCAUUGGC--CUUAUGUGUCGACUCCGGCAUUCGGCUGAGAUAGGGCU (((((((((((((((((((.((....))))))...(((((-(..((((((.......))---).)))...))--)))))))))))....((((.....)))).)))))))). ( -43.20) >DroEre_CAF1 9857 106 - 1 GCCUUAUCCGGCACAAACCAGUUGGUUCGAUUCCUUGACG-CGCUGCUCAAGUCCACAG---GUCCACUGGC--UUUAUUUGCCGACACUGGCAUUCGGCUGAGAUAGGGCU ((((((((((((.....(((((.((..(.....(((((.(-(...)))))))......)---..))))))).--......(((((....)))))....)))).)))))))). ( -38.50) >DroYak_CAF1 4264 106 - 1 GCCUUGUCCGGCACAAACCAGUUGGUGCGGUUCCUUGAGG-CACUGGCCAAGUCCACUG---GCCCACUUGC--CUUAUUUGCCGACUGCGGCAUUCGGCUGAGAUAGGGCU ((((((((((((....(((....))).(((.....(((((-((..(((((.......))---)))....)))--))))..(((((....))))).))))))).)))))))). ( -45.90) >DroMoj_CAF1 5962 106 - 1 GCCUUGUCGCCCACGAACCAAUUGGUGCGAUUGCGCGACGGCGCU-GCCAGGUCCACAGUUGCCUGGUUGGCCGC-----UGGCUUCUCCUCCACACGACUGAGAUAGGGCC (((((((((((..((..(((((..(.(((((((.(.((((((...-)))..)))).))))))))..))))).)).-----.)))..........((....)).)))))))). ( -41.30) >DroPer_CAF1 16208 99 - 1 ----CGUCUGCCACAAACCAAUUGGUGCGAUUUCUUGAGG-CGCUGGCCAAGUCCACCAUGGAUCUGUUGGC--------GGCCCCCUCGGGUAUCUUGCUCAGAUAGGGCC ----.....((((((..(((..(((((.(((((.....((-(....))))))))))))))))...)).))))--------(((((..((((((.....)))).))..))))) ( -36.30) >consensus GCCUUAUCCGGCACAAACCAAUUGGUGCGAUUCCUUGAGG_CACUGGCCAAGUCCACUG___GUCCACUGGC__CUUAUUUGCCGACUCCGGCAUUCGGCUGAGAUAGGGCU ((((((((.........((((((((((.((...((((...........))))))))))).......))))).........(((((....))))).........)))))))). (-18.97 = -19.36 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:19 2006